Tuesday, March 20, 2018 From rOpenSci (https://ropensci.org/blog/2018/03/20/rentrez-paper/). Except where otherwise noted, content on this site is licensed under the CC-BY license.
I am happy to say that the latest issue of The R Journal includes a paper describing rentrez, the rOpenSci package for retrieving data from the National Center for Biotechnology Information (NCBI).
The NCBI is one of the most important sources of biological data. The centre
provides access to information on 28 million scholarly articles through PubMed and 250
million DNA sequences through GenBank. More importantly, records in the 50 public
databases maintained by the NCBI are strongly cross-referenced. As a result, it is
possible to pinpoint searches using almost 2 million taxonomic names or a
controlled vocabulary with 270,000 terms.
rentrez has been designed to make it easy to search for and download NCBI
records and download them from within an R session.
The paper and the package vignette
both describe typical usages of rentrez. I though it might be fun to use this
post to find out where papers describing R packages are published these days.
Although PubMed only covers journals in the biological sciences, searching that
database will at least give us an idea of which journals like to publish these
sorts of papers. Here we use the entrez_search and entrez_summary functions
to get some information on all of the papers published in 2017 with the term
‘R package’ in their title:
library(rentrez)
pkg_search <- entrez_search(db="pubmed",
term="(R Package[TITLE]) AND (2017[PDAT])",
use_history=TRUE)
pkg_summs <- entrez_summary(db="pubmed", web_history=pkg_search$web_history)
pkg_summs
List of 96 esummary records. First record:
$`29512507`
esummary result with 42 items:
[1] uid pubdate epubdate source
[5] authors lastauthor title sorttitle
[9] volume issue pages lang
[13] nlmuniqueid issn essn pubtype
[17] recordstatus pubstatus articleids history
[21] references attributes pmcrefcount fulljournalname
[25] elocationid doctype srccontriblist booktitle
[29] medium edition publisherlocation publishername
[33] srcdate reportnumber availablefromurl locationlabel
[37] doccontriblist docdate bookname chapter
[41] sortpubdate sortfirstauthor
As you can tell from the output above, you can get a lot of information from
these summary records. In this case, we are interested in the journals in which
these papers appear. We can use the helper function extract_from_esummary
to isolate the ‘source’ of each paper, then use table to count up the frequency
of each journal.
library(ggplot2)
journals <- extract_from_esummary(pkg_summs, "source")
journal_freq <- as.data.frame(table(journals, dnn="journal"), responseName="n.papers")
ggplot(journal_freq, aes(reorder(journal, n.papers), n.papers)) +
geom_point(size=2) +
coord_flip() +
scale_y_continuous("Number of papers") +
scale_x_discrete("Journal") +
theme_bw() +
ggtitle("Venues for papers describing R Packages in 2017")
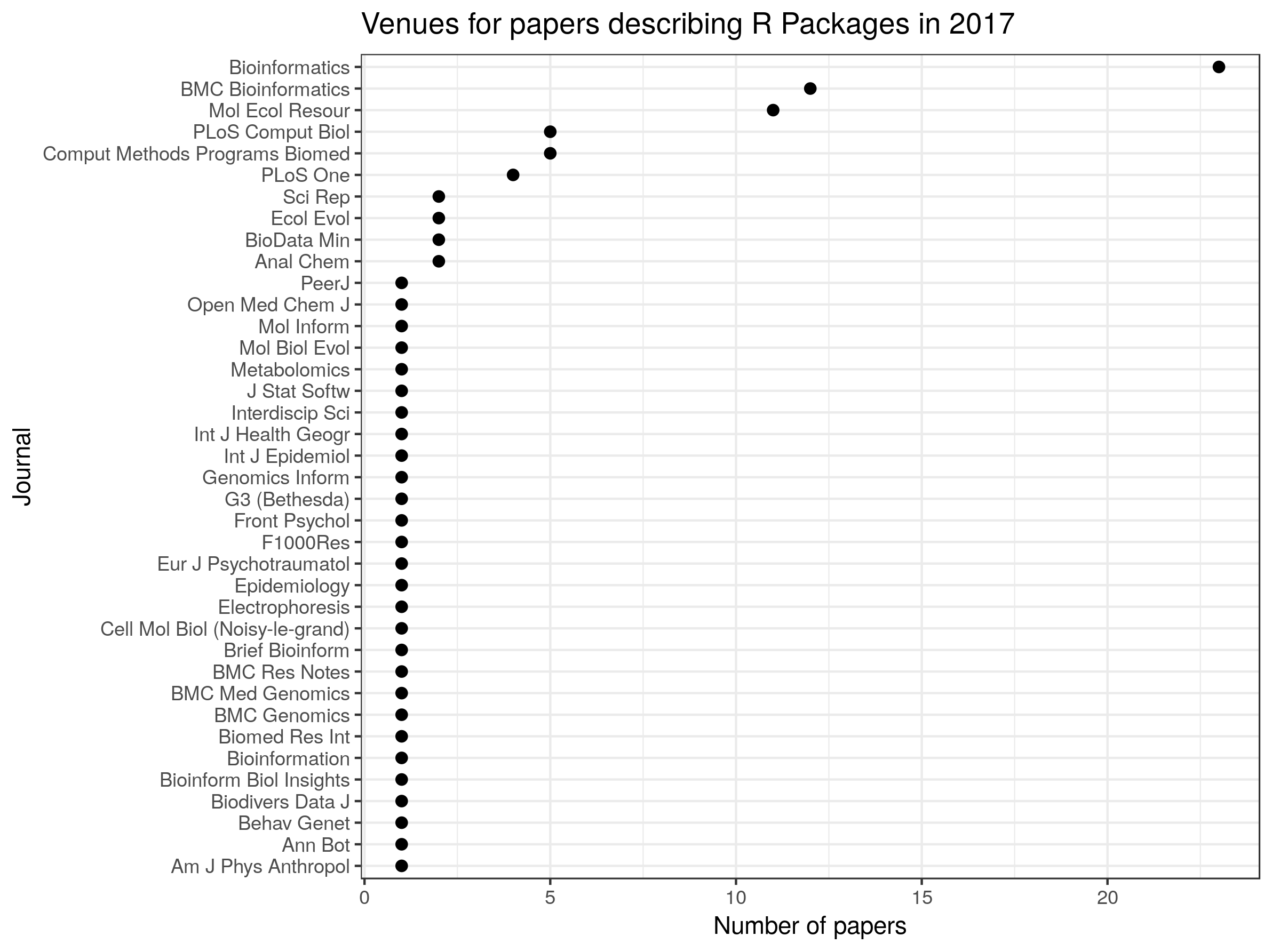
So, it looks like Bioinformatics, BMC Bionformatics and Molecular Ecology Resources are popular destinations for papers describing R packages, but these appear in journals all the way across the biological sciences.
The R Journal article describes some more typical uses of rentrez, and also
describes some of decisions that went into the design of the package. If this
example has whetted your appetite, then please check out the article or the
package documentation.
🔗 Thanks
The publication of this paper gives me a chance to thank the
many people that have helped make rentrez into a useful package. I was very
lucky to have this code included in rOpenSci at an early stage. Being part of
the wider project made sure rentrez kept pace with the best-practices for code
and documentation developed by the R community and got the package out to a wider
audience than would have otherwise been possible. I am thankful to everyone who has
filed an issue or contributed code to rentrez. I also have to
single out Scott Chamberlain, who has done a great deal to make sure the code
meets community standards and is useful to as many people as possible.
🔗 API keys for eUtils
To celebrate the publication of this paper I am going to speed up rentrez by a
factor of three!
Well, the timing is coincidental, but the latest release of rentrez does make it
possible to send and receive information from the NCBI at a greater rate than
was previously possible. The NCBI now gives users the opportunity to register for an access
key
that will allow them to make up to 10 requests per second (non-registered users are limited
to 3 requests per second per IP address). As of the latest release, rentrez
supports the use of these access keys while enforcing the appropriate rate limits.
For one-off cases, this is as simple as adding the api_key argument to a given
function call.
prot_links <- entrez_link(db="protein", dbfrom="gene", id=93100, api_key ="ABCD123")
It most cases you will want to use your key for each of several calls to the
NCBI. rentrez makes this easy by allowing you to set an environment variable,
ENTREZ_KEY. Once this value is set to your key rentrez will use it for all
requests to the NCBI. To set the value for a single R session you can use the
function set_entrez_key(). Here we set the value and confirm it is now
available as an environment variable.
set_entrez_key("ABCD123")
Sys.getenv("ENTREZ_KEY")
## [1] "ABCD123"
If you use rentrez often you should edit your .Renviron file (see
help(Startup) for a description of this file) to include your key. Doing so will
mean all requests you send will take advantage of your API key. Here’s the line
to add:
ENTREZ_KEY=ABCD123
As long as an API key is set by one of these methods, rentrez will allow you
to make up to ten requests per second.
🔗 Bugs and use-cases please!
The publication in the R Journal is not the end of development for rentrez.
Though the package is now feature-complete and stable, I am very keen to make sure
it keeps pace with the API it wraps and squash any bugs that might arise. I also
appreciate use-cases that demonstrate how the package can take advantage of NCBI
data. So, please, file issues at the project’s repository if you have any
questions about it!

