Tuesday, August 14, 2018 From rOpenSci (https://ropensci.org/blog/2018/08/14/where-to-bird/). Except where otherwise noted, content on this site is licensed under the CC-BY license.
The blog post series corresponds to the material for a talk Maëlle will give at the Animal Movement Analysis summer school in Radolfzell, Germany on September the 12th, in a Max Planck Institute of Ornithology.
This post is the 1st post of a series showcasing various rOpenSci packages as if Maëlle were a birder trying to make the most of R in general and rOpenSci in particular. Although the series use cases will mostly feature birds, it’ll be the occasion to highlight rOpenSci’s packages that are more widely applicable, so read on no matter what your field is! Moreoever, each post should stand on its own.
A logical first step in this birder’s guide to rOpenSci is to use R to find where to observe birds! In this blog post, we shall use rOpenSci packages accessing open geographical data to locate and visualize where to observe birds near a given location.
🔗 First of all, where are we now?
The Max Planck Institute for Ornithology (henceforth shortened to MPI),
where Maëlle will give a talk is located at Am Obstberg 1 78315
Radolfzell. Let’s geolocate it using rOpenSci’s opencage
package that interfaces the
OpenCage Geocoder, a commercial service
based on open data. When choosing to
get only one result via limit = 1, one gets what the API considers to
be the best one.
mpi <- opencage::opencage_forward("Am Obstberg 1 78315 Radolfzell", limit = 1)$results
class(mpi)
## [1] "tbl_df" "tbl" "data.frame"
head(names(mpi))
## [1] "annotations.DMS.lat" "annotations.DMS.lng"
## [3] "annotations.MGRS" "annotations.Maidenhead"
## [5] "annotations.Mercator.x" "annotations.Mercator.y"
This gets us Am Obstberg 1, 78315 Radolfzell, Germany (mpi$formatted)
which is in 🇩🇪 (mpi$annotations.flag gets us a flag!).
🔗 Birding in a bird hide?
🔗 Where to find a bird hide?
You can most certainly go birding anywhere, but if you can find a bird
hide (or bird blind depending on the English you speak), it might be a
very appropriate observation point. Now that we know where the MPI is,
we can look for bird hide(s) in the vicinity. For that, we shall use
rOpenSci’s osmdata package by
Mark Padgham and collaborators! Note that incidentally, Mark did his
PhD in
ecology.
This package is an interface to OpenStreetMap’s Overpass
API. OpenStreetMap is
a collective map of the world. It contains information about towns’
limits, roads, placenames… but also tags of everything, from bars as
seen in this blog post to
trees. You can
browse existing features via OpenStreetMap’s
wiki. Some parts of the
world are better mapped than others depending on the local OpenStreetMap
community. Actually, OpenCage’s blog features an interesting series of
country profiles.
To look for a bird hide, we first create a bounding box of 10km around
the MPI, using rOpenSci’s bbox
package.
bbox <- bbox::lonlat2bbox(mpi$geometry.lng,
mpi$geometry.lat,
dist = 10, method = "lawn")
We then use the key and value associated with bird hides in OpenStreetMap: respectively leisure and bird_hide.
library("osmdata")
## Data (c) OpenStreetMap contributors, ODbL 1.0. http://www.openstreetmap.org/copyright
library("magrittr")
(results <- opq(bbox = bbox) %>%
add_osm_feature(key = 'leisure', value = 'bird_hide') %>%
osmdata_sf ())
## Object of class 'osmdata' with:
## $bbox : 47.6865,8.8753,47.8494,9.1177
## $overpass_call : The call submitted to the overpass API
## $timestamp : [ Thu 5 Jul 2018 08:06:35 ]
## $osm_points : 'sf' Simple Features Collection with 1 points
## $osm_lines : 'sf' Simple Features Collection with 0 linestrings
## $osm_polygons : 'sf' Simple Features Collection with 0 polygons
## $osm_multilines : 'sf' Simple Features Collection with 0 multilinestrings
## $osm_multipolygons : 'sf' Simple Features Collection with 0 multipolygons
results$osm_points
## leisure geometry
## 5004940425 bird_hide 8.920901, 47.741569
Yay, we now know where to find a bird hide not too far from the MPI!
🔗 Visualizing our location and the bird hide
So one could enter the coordinates of that bird hide in one’s favourite
mapping software or app but to show you where the bird hide is we can
actually step back and use
osmplotr, another package
contributed to rOpenSci by Mark Padgham!
The way osmplotr works is letting you create a basemap, on which
you’ll add different layers extracted from OpenStreetMap using
osmplotr::extract_osm_objects or osmdata functions directly. Its
strengths are therefore the use of open data, and the customization of
what you’re using as background!
Let’s create a basemap for our bounding box, and then add roads and buildings to it.
library("osmplotr")
## Data (c) OpenStreetMap contributors, ODbL 1.0. http://www.openstreetmap.org/copyright
bbox <- get_bbox(bbox)
dat_B <- extract_osm_objects (key = 'building', bbox = bbox)
dat_H <- extract_osm_objects (key = 'highway', bbox = bbox)
map0 <- osm_basemap(bbox = bbox, bg = 'gray20') %>%
add_osm_objects (dat_B, col = 'gray40') %>%
add_osm_objects (dat_H, col = 'gray80')
map0 %>%
add_axes() %>%
print_osm_map (filename = 'map_a1.png', width = 600,
units = 'px', dpi = 72)
library("magrittr")
magick::image_read('map_a1.png') %>%
magick::image_annotate("Map data © OpenStreetMap contributors",
color = "white",
boxcolor = "black",
size = 15,
gravity = "south")
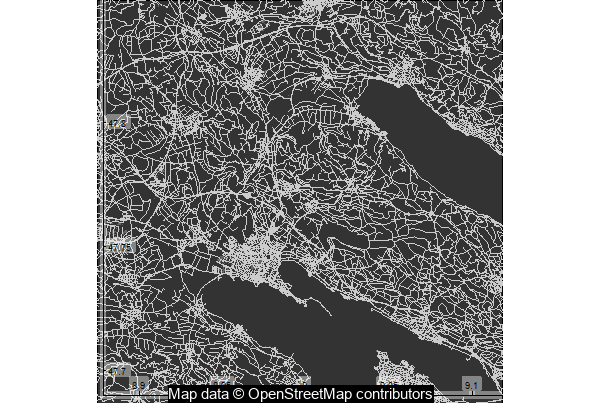
Quite pretty! The lakes can be seen because of the absence of roads and buildings on them.
Now, let’s plot the bird hide and the MPI on them. Since we used
osmdata::osmdata_sf, we had gotten the data in a receivable class,
sf.
points_map <- add_osm_objects(map0, results$osm_points,
col = 'salmon',
size = 5)
For plotting the MPI, we’ll convert opencage output to an sf point
with the same coordinate reference system as the OpenStreetMap data
extracted with osmdata.
coords <- data.frame(lon = mpi$geometry.lng,
lat = mpi$geometry.lat)
crs <- sf::st_crs(results$osm_points)
mpi_sf <- sf::st_as_sf(coords,
coords = c("lon", "lat"),
crs = crs)
points_map <- add_osm_objects(points_map, mpi_sf,
col = 'white',
size = 5)
We can now visualize both points on the map, the MPI in white and the bird hide in salmon, South-West from the MPI.
points_map %>%
add_axes() %>%
print_osm_map (filename = 'map_a2.png',
width = 600,
units = 'px', dpi = 72)
magick::image_read('map_a2.png') %>%
magick::image_annotate("Map data © OpenStreetMap contributors",
color = "white",
boxcolor = "black",
size = 15,
gravity = "south")

Aha, now we see where the bird hide is, fantastic! But as Mark noted, birds can actually be observed from other places.
🔗 Birding where birds should be?
Birds are most likely to be found where water lies close to natural
areas, and we can translate this to R code! We shall get all water and
(separately) all non-watery natural areas and find the shortest
distances between them before plotting the results using
add_osm_surface.
First, let’s get all water and (separately) all non-watery natural areas.
dat <- opq(bbox = bbox) %>%
add_osm_feature(key = 'natural') %>%
osmdata_sf (quiet = FALSE)
## Issuing query to Overpass API ...
## Rate limit: 2
## Query complete!
## converting OSM data to sf format
indx_W <- which (dat$osm_polygons$natural %in% c ("water", "wetland"))
indx_N <- which (!dat$osm_polygons$natural %in% c ("water", "wetland"))
xy_W <- sf::st_coordinates (dat$osm_polygons [indx_W, ])
xy_N <- sf::st_coordinates (dat$osm_polygons [indx_N, ])
Then we use Mark’s geodist
package to get pairwise distances
between all points, find minima, and make a data.frame to submit to
add_osm_surface(). We have 7068 watery points and 10065 non-watery
points so the speed of geodist is crucial here!
t1 <- Sys.time()
d <- geodist::geodist (xy_W, xy_N)
# so fast!!!
Sys.time() - t1
## Time difference of 13.57983 secs
d1 <- apply (d, 1, min)
d2 <- apply (d, 2, min)
xy <- cbind (rbind (xy_W, xy_N), c (d1, d2))
xy <- xy [, c (1, 2, 5)]
colnames (xy) <- c ("x", "y", "z")
xy <- xy [!duplicated (xy), ]
Finally we plot the results on the roads we had gotten earlier, although
we do not recommend staying on the middle of a road to observe birds! We
re-add the points corresponding to the MPI and bird hide after the
surface coloring. With osmplotr, order matters because layers are
added on top of each other. Note that plotting the distances takes a
while.
# colorblind-friendly palette!
cols <- viridis::viridis_pal (direction = -1) (30)
add_osm_surface (map0, dat_H,
dat = xy, col = cols) %>%
add_axes() %>%
add_colourbar(cols = cols,
zlim = range(xy[,"z"])) %>%
add_osm_objects(mpi_sf,
col = 'white', size = 5) %>%
add_osm_objects(results$osm_points,
col = 'white', size = 5)%>%
print_osm_map (filename = 'map_a3.png', width = 600,
units = 'px', dpi = 72)
magick::image_read('map_a3.png') %>%
magick::image_annotate("Map data © OpenStreetMap contributors",
color = "white",
boxcolor = "black",
size = 15,
gravity = "south")

On the map, the yellower/lighter a road is, the better it is to observe birds according to Mark’s assumption that birds are most likely to be found where water lies close to natural areas. With this metric, the MPI itself is not too badly located, which after all is not too surprising for an MPI of ornithology. Maybe one should just walk to the one of the nearest lakes to meet some birds.
🔗 Conclusion
🔗 Open geographical data in R
In this post we showcased three rOpenSci packages helping you use open
geographical data in R:
opencage,
osmdata,
osmplotr, therefore mostly
making use of the awesome OpenStreetMap data (The OpenCage Geocoder uses
a bit more, but it only warrants attributing
OpenStreetMap). We therefore added
the annotation “Map data © OpenStreetMap contributors” to all maps of
this post using rOpenSci’s magick package. You might also have noticed
in the code earlier that both osmdata and osmplotr have start-up
messages indicating the data origin and licence.
Another package of rOpenSci’s interacting with open geographical data,
that might be of interest for making maps, is
rnaturalearth that
facilitates interaction with Natural Earth map
data.
🔗 Other R packages for spatial analyses
We also used two other rOpenSci packages:
bbox to get a bounding box and
magick for image manipulation.
Explore more of our packages suite, including and beyond the geospatial
category, here.
We also used the geodist
package for ultra lightweight,
ultra fast calculation of geo distances. This package is developed in
the hypertidy GitHub organization which
is a good place to find useful R geospatial packages. Other good
resources for geospatial analyses with R include the r-spatial.org
website and this great book by Robin
Lovelace, Jakub Nowosad and Jannes
Muenchow, and more links
presented in this blog post of Steph
Locke’s.
🔗 More birding soon!
Stay tuned for the next post in this series, about getting bird observation data in R! In the meantime, happy birding!

