Monday, October 8, 2018 From rOpenSci (https://ropensci.org/blog/2018/10/08/orcid/). Except where otherwise noted, content on this site is licensed under the CC-BY license.
Proper identification of individuals is crucial for acknowledging and studying their scientific work, be it journal articles or pieces of software. In this tech note, one year after CRAN started supporting ORCIDs, we shall explain why and how to use unique author identifiers in DESCRIPTION files.
🔗 Why use ORCIDs on CRAN?
When analyzing the authorship of CRAN packages, one can look at authors’ names and email addresses. Names can be written with and without quotes, email addresses change, which makes it all tricky as noted by David Smith when he looked for the most prolific CRAN authors (notice our very own Scott Chamberlain and Jeroen Ooms in that scoreboard by the way?). Besides, several people can have the same name!
So all in all, using an unique ID per author makes sense. That’s something offered for the academic world and beyond by ORCID, ORCID meaning “Open Researcher and Contributor ID”. Anyone can set up an ORCID account for free and link it up to the rest of their online persona, as well as to their employment and works. When the authors of a scientific paper give their ORCID ID to the publisher, these can be included in the html and PDF versions of a paper. Note that there’s no, say, Keybase verification for ORCIDs, and often the input of an ORCID ID for a paper is declarative as well. Read more about ORCID here.
About one year ago, CRAN added support for
ORCID in
DESCRIPTION files by a minimally invasive
hack. One
can add the ORCID ID of an author as a comment field in person(),
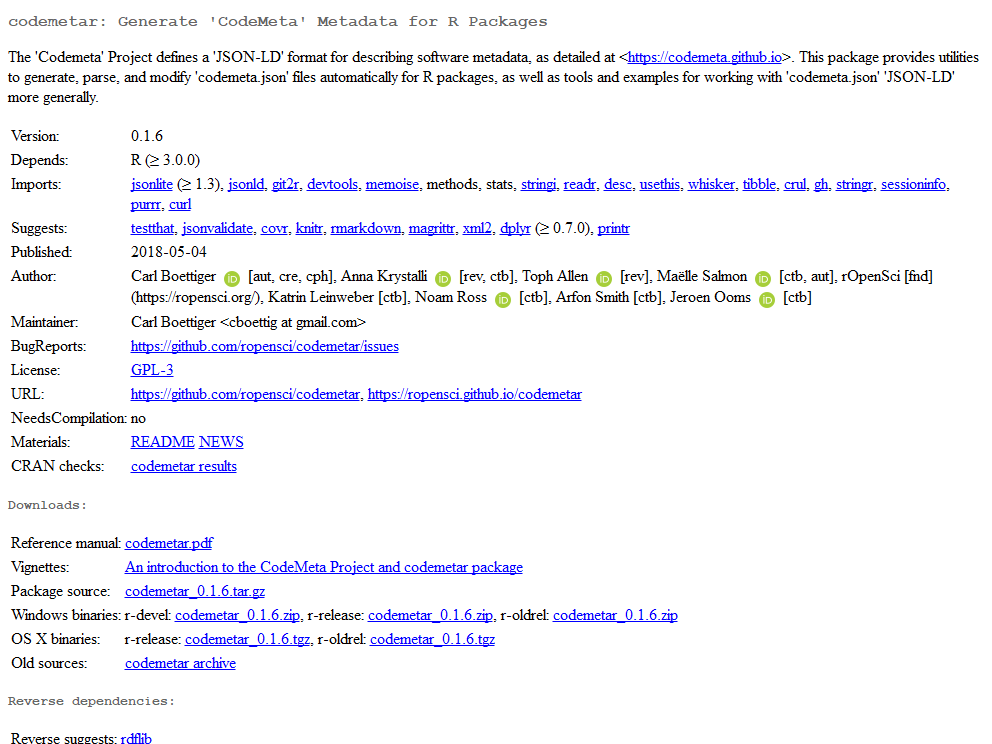
see e.g. Carl Boettiger’s ID in codemetar
DESCRIPTION,
and one gets a nice little green bubble near their name on CRAN, see
e.g. codemetar CRAN
page.
The advantages of using ORCID IDs in DESCRIPTION are:
Unique identifiers for free! Sure, there’s no verification so someone could use your identity, but at least, it should help assigning your own work to you.
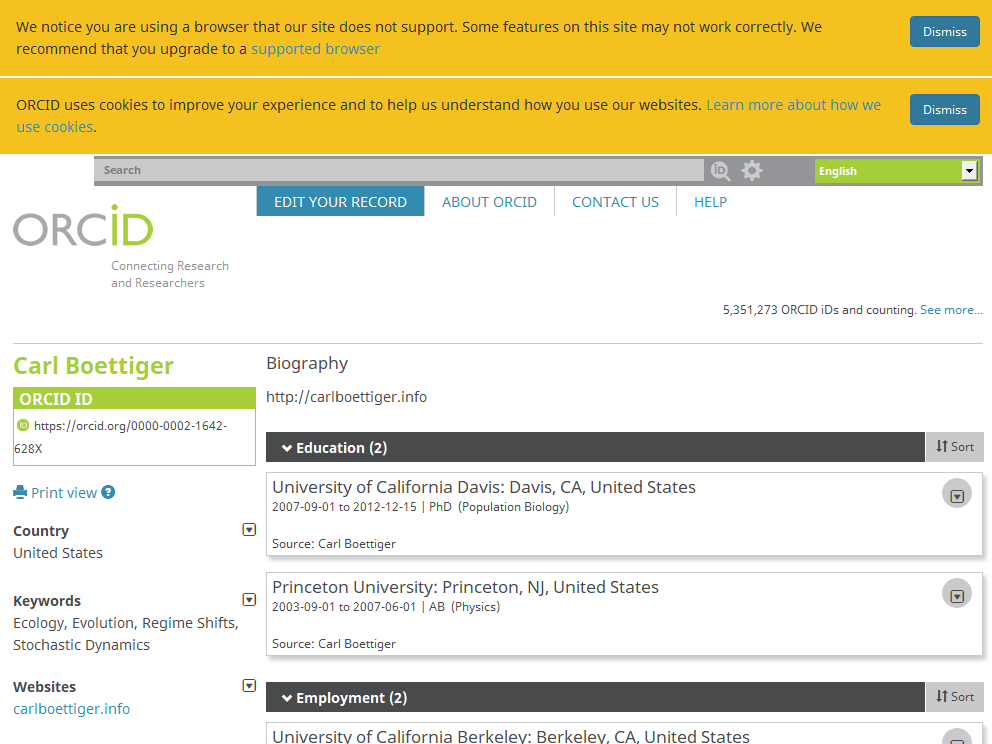
Clickable name on CRAN pages, METACRAN pages and pkgdown websites! E.g. when clicking on Carl’s green bubble on
codemetarCRAN page, one gets to his ORCID profile including a link to his personal website. See alsocodemetarMETACRAN page andcodemetarpkgdown website.
Even for non academics, making one’s online persona more accessible from a CRAN DESCRIPTION can’t hurt.
🔗 How to manipulate ORCIDs in DESCRIPTION?
So, how does one add add and interact with ORCID in DESCRIPTION? Well,
thankfully, the current dev version of the RStudio desc
package by Gábor
Csárdi has some direct support of
ORCIDs! Gábor too is well ranked in the scoreboard mentioned
earlier!
remotes::install_github("r-lib/desc")
🔗 Search authors by ORCID
desc offers many functions for manipulating authors: adding or
deleting them or their roles. Now you can perform these operations using
ORCID to identify authors to be modified. E.g. if you want to add a role
to an existing author, say Carl who worked hard enough to earn the “aut”
role, you’d run
desc::desc_add_role(role = "aut",
orcid = "0000-0002-1642-628X")
Obviously this very example above has probably little real-life applicability, but you could imagine manipulating DESCRIPTION files en masse using a table of ORCIDs and new roles.
🔗 Add ORCID to authors of a package
The new add_orcid() method and desc_add_orcid() functions make it
possible to add ORCID IDs to authors directly instead of via the
comment argument. So you could write some script
desc::desc_add_orcid("0000-0002-2815-0399",
given = "Maëlle",
family = "Salmon")
and run it inside all your local package folders to add your ORCID. It will add this ORCID ID to the author whose given name is “Maëlle” and whose family name is “Salmon”. If you only write
desc::desc_add_orcid("0000-0002-2815-0399",
given = "Maëlle")
and there are two Maëlle’s, the function will error and you’ll need to run it with more specific arguments.
🔗 Add all of your identity at once!
The coolest feature of desc ORCID support might be that
desc::desc_add_me() will now use the ORCID of the environment variable
ORCID_ID if you created it. Hence, once per machine it’d now be
recommended to
install the
whoamipackage necessary fordesc::desc_add_me()to work.install.packages("whoami").create an environment variable
ORCID_ID, editing.Renvironby calling e.g.usethis::edit_r_environ(). Here’s how the corresponding.Renvironline looks for me:
ORCID_ID="0000-0002-2815-0399"
desc::desc_add_me() looks for your name and email addresses in your
existing configuration (git configuration, GitHub profile), see the
whoami package docs. So say you’ve
just been told to add yourself as contributor of a package you’ve
improved: inside the package directory, you’d just need to run
desc::desc_add_me() and voilà ! No risk to wrongly copy-paste your
ORCID.
As a conclusion of this section, not only are ORCIDs useful, but they’re getting citizen status in the world of as-automatic-as-possible package development. Now, we shall see how one could use ORCIDs inside R to gather information about package authors.
🔗 How to study CRAN authors via their ORCID
As mentioned earlier, rOpenSci’s Scott Chamberlain is the most prolific
CRAN package
maintainer,
so perhaps unsurprisingly, he maintains a package interacting with
ORCID’s API, rorcid!
The API requires authentication so the first step after installing the package is to run:
rorcid::orcid_auth()
and to save the resulting token as an ORCID_TOKEN environment variable
in .Renviron (again, usethis::edit_r_environ() can help with that).
Authentication warrants your having an ORCID account, so if you haven’t
one yet, register via this link.
Then, we’ll query all comment’s of Authors@R of CRAN packages. Some
CRAN packages use the Author field instead, which is not recommended
and doesn’t allow using comment.
Side-note, in general, how would one analyze stuff over the whole CRAN collection?
Some information can be found in
tools::CRAN_package_db()as seen in this recent tech note.There’s an official read-only mirror of each all CRAN packages on GitHub thanks to METACRAN.
METACRAN serves a CRAN package database, wrapped by an R package of course.
One could try setting up a local CRAN mirror.
Thanks to Gábor, who maintains METACRAN, for patiently answering my questions about meta-CRAN things. If we were after the comments section of just a few packages, we could query METACRAN database like so:
url <- "https://crandb.r-pkg.org/-/latest?keys=[%22desc%22,%22codemetar%22]"
obj <- jsonlite::fromJSON(url, simplifyVector = FALSE)
lapply(obj, function(x) eval(parse(text = x$`Authors@R`))$comment)
but in our case, when wanting to get all the current CRAN authors, it’s
easier to use tools::CRAN_package_db(). As a side-note, if you find a
better solution than the functions defined below parse_authors() and
parse_author() (some purrr magic?) to coerce all authors information
to a data.frame, feel free to comment under the post!
library("magrittr")
all_info <- tools::CRAN_package_db()
all_info <- all_info[!is.na(all_info$`Authors@R`),]
all_info <- all_info[, c("Package", "Authors@R")]
names(all_info)[2] <- "authors_at_r"
null_to_na <- function(x){
if(is.null(x)){
NA
}else{
x
}
}
parse_author <- function(author){
author <- as.person(author)
tibble::tibble(given = null_to_na(author$given),
family = null_to_na(author$family),
email = null_to_na(author$email),
comment = as.character(null_to_na(author$comment)),
orcid = as.character(null_to_na(author$comment["ORCID"])))
}
parse_authors <- function(authors_at_r){
authors <- try(eval(parse(text = authors_at_r)),
silent = TRUE)
if(inherits(authors, "try-error")){
return(list(NA))
}
else{
list(purrr::map_dfr(authors, parse_author))
}
}
cran_authors <- all_info %>%
as.data.frame() %>%
dplyr::select(Package, authors_at_r) %>%
dplyr::rowwise() %>%
dplyr::mutate(authors = parse_authors(authors_at_r)) %>%
tidyr::unnest(authors)
This is how the resulting table looks like. Out of 13147 packages on
CRAN we are looking at 5421, i.e. packages using the Authors@R field.
There is one line per author in a package.
str(cran_authors)
## Classes 'tbl_df', 'tbl' and 'data.frame': 16272 obs. of 7 variables:
## $ Package : chr "abbyyR" "abc" "abc" "abc" ...
## $ authors_at_r: chr "person(\"Gaurav\", \"Sood\", email = \"[email protected]\", role = c(\"aut\", \"cre\"))" "c( \n person(\"Csillery\", \"Katalin\", role = \"aut\", email=\"[email protected]\"),\n person(\"Le"| __truncated__ "c( \n person(\"Csillery\", \"Katalin\", role = \"aut\", email=\"[email protected]\"),\n person(\"Le"| __truncated__ "c( \n person(\"Csillery\", \"Katalin\", role = \"aut\", email=\"[email protected]\"),\n person(\"Le"| __truncated__ ...
## $ given : chr "Gaurav" "Csillery" "Lemaire" "Francois" ...
## $ family : chr "Sood" "Katalin" "Louisiane" "Olivier" ...
## $ email : chr "[email protected]" "[email protected]" NA NA ...
## $ comment : chr NA NA NA NA ...
## $ orcid : chr NA NA NA NA ...
We can count the number of occurrences of ORCIDs.
(cran_authors %>%
dplyr::filter(!is.na(orcid)) %>%
dplyr::pull(Package) %>%
unique() %>%
length() -> orcid_pkg_no)
## [1] 613
Only 613 packages have at least an ORCID ID out of 13147 CRAN packages
(you can’t have an ORCID ID without using the Authors@R field).
There are 512 unique ORCID IDs. Let’s look at the most prolific authors with an ORCID ID.
cran_authors %>%
dplyr::filter(!is.na(orcid)) %>%
dplyr::count(orcid, sort = TRUE) %>%
head(n = 10) -> most_prolific
knitr::kable(most_prolific)
| orcid | n |
|---|---|
| 0000-0003-1444-9135 | 29 |
| 0000-0002-4035-0289 | 21 |
| 0000-0003-0918-3766 | 21 |
| 0000-0003-4198-9911 | 19 |
| 0000-0003-4097-6326 | 16 |
| 0000-0001-8301-0471 | 13 |
| 0000-0003-0645-5666 | 12 |
| 0000-0001-5243-233X | 11 |
| 0000-0001-5670-2640 | 11 |
| 0000-0002-8584-459X | 10 |
Then we can use rorcid to get a glimpse of their profile.
rorcid::as.orcid(most_prolific$orcid)
## [[1]]
## <ORCID> 0000-0003-1444-9135
## Name: Chamberlain, Scott
## URL (first): /
## Country: US
## Keywords: ecology, open access, bioinformatics, evolution, R
##
## [[2]]
## <ORCID> 0000-0002-4035-0289
## Name: Ooms, Jeroen
## URL (first): https://github.com/jeroen
## Country: NL
## Country: US
## Keywords:
##
## [[3]]
## <ORCID> 0000-0003-0918-3766
## Name: Zeileis, Achim
## URL (first): https://eeecon.uibk.ac.at/~zeileis/
## Country: AT
## Keywords:
##
## [[4]]
## <ORCID> 0000-0003-4198-9911
## Name: Hornik, Kurt
## URL (first):
## Country:
## Keywords:
##
## [[5]]
## <ORCID> 0000-0003-4097-6326
## Name: Leeper, Thomas
## URL (first): http://www.lse.ac.uk/government/whosWho/Academic%20profiles/ThomasLeeper.aspx
## Country: GB
## Keywords: Public Opinion, Survey Experiments, Political Behaviour, R, Research Design
##
## [[6]]
## <ORCID> 0000-0001-8301-0471
## Name: Hothorn, Torsten
## URL (first):
## Country:
## Keywords:
##
## [[7]]
## <ORCID> 0000-0003-0645-5666
## Name: Xie, Yihui
## URL (first): https://yihui.name
## Country:
## Keywords:
##
## [[8]]
## <ORCID> 0000-0001-5243-233X
## Name: Tang, Yuan
## URL (first): https://terrytangyuan.github.io/about/
## Country: US
## Keywords: Machine Learning, Data Visualization, Open Source, Software Engineering
##
## [[9]]
## <ORCID> 0000-0001-5670-2640
## Name: Rudis, Bob
## URL (first): http://rud.is/b
## Country: US
## Keywords: cybersecurity, r, data visualization, web crawling/scraping
##
## [[10]]
## <ORCID> 0000-0002-8584-459X
## Name: You, Kisung
## URL (first): https://kisungyou.github.io/
## Country: KR
## Keywords:
Interestingly we recognize some names from the most prolific CRAN maintainers (Scott and Jeroen, again!) but not only. Note that another difference with David Smith’s post is that the list above includes authors no matter their role.
Over all authors with an ORCID, using rorcid one could extract their
location and much more, but this is beyond the scope of this tech note.
Inversely, if one wanted to find CRAN packages by ORCID ID, instead of ORCID IDs by CRAN package, one could make the most of METACRAN. There is no search by ORCID, but since everything in DESCRIPTION is indexed by METACRAN, one can use the ORCID ID as search term. Say you want to find all packages by Scott Chamberlain:
You can use this URL https://r-pkg.org/search.html?q=0000-0003-1444-9135
Or this to get an API (and further parse the JSON in R?) http://seer.r-pkg.org:9200/cran-devel/package/_search?q=%220000-0003-1444-9135%22
Other methods to search CRAN by ORCID ID might include using the
seerpackage (not possible yet).
🔗 Conclusion
In this tech note we made the case for using ORCID IDs as identifiers
for the authors of CRAN packages, for academics and non-academics as
well. We described the current desc
support for ORCID. We also gave a small insight into the wealth of
information one can get via
rorcid.
For academics, another aspect of making their software contributions valuable is getting the software they write as work inside their ORCID profile. One can do that by hand, but one can also
get a legit DOI for each package release by activating Zenodo for the repository,
write a software paper, e.g. via JOSS, that’d have its own DOI.
But this is probably a topic for another time, such as would be the topic of CRAN potentially adopting other IDs like GitHub username, ideally via Keybase. By the way if you’d like to see a Keybase integration of ORCID happen, please “+1” this issue by Steph Locke. In the meantime, let’s hope ORCID IDs will be more adopted by academics and non-academics alike for:
Making CRAN package authors easier to identify and their online persona easier to find.
Making academic CRAN package authors’ scientific contributions easier to access when studying packages.
Have you started desc::add_orcid()-ing? Don’t hesitate to comment
below!
Many thanks to 0000-0002-1642-628X and 0000-0003-1419-2405 for their feedback on this note.

