Thursday, April 18, 2019 From rOpenSci (https://ropensci.org/blog/2019/04/18/wild-standards/). Except where otherwise noted, content on this site is licensed under the CC-BY license.
This post is published on the rOpenSci and Methods in Ecology and Evolution blogs
🔗
 Stefanie Butland, rOpenSci Community Manager
Stefanie Butland, rOpenSci Community Manager
Some things are just irresistible to a community manager – PhD student Hugo Gruson’s recent tweets definitely fall into that category.
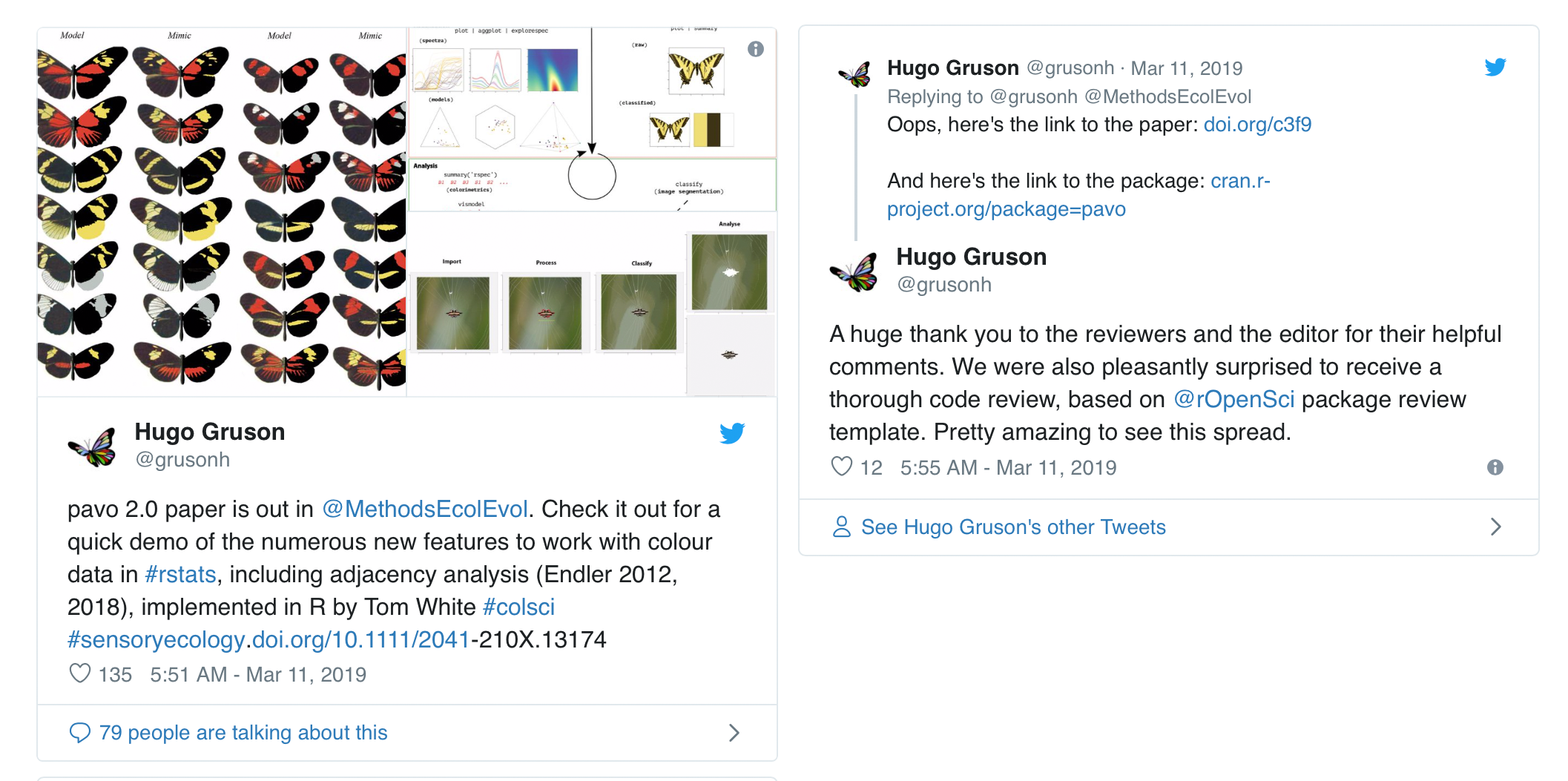
I was surprised and intrigued to see an example of our software peer review guidelines being used in a manuscript review, independent of our formal collaboration with the journal Methods in Ecology and Evolution (MEE). This is exactly the kind of thing rOpenSci is working to enable by developing a good set of practices that broadly apply to research software.
But who was this reviewer and what was their motivation? What role did the editors handling the manuscript play? I contacted the authors and then the journal and, in less than a week we had everyone on board to talk about their perspectives on the process.
🔗
 Nick Golding, Associate Editor, Methods in Ecology and Evolution
Nick Golding, Associate Editor, Methods in Ecology and Evolution
To me, MEE’s role is to help increase the quality of the methods used in ecology and evolution, and this includes research software. It would be great to reach a point where all the research software used in ecology is at the same high standard as the packages that have been through rOpenSci software peer review.
Not all R packages that we receive at MEE fit in with the rOpenSci package scope, but I’d love to see them go through a similar process. This is where the rOpenSci review checklist comes in. In my view, it’s the gold standard 🏆 for reviewing R packages and I was thrilled to see that Hao (manuscript reviewer) had used it with this paper.
The idea of doing code review as part of reviewing a manuscript is new to a lot of people. Often, invited reviewers decline because they don’t think they have the right experience. If you have experience with creating packages though, reviewing code isn’t something to be worried about. rOpenSci’s guidelines are a great way for people new to reviewing code to become comfortable with the process.
🔗
 Hao Ye, Manuscript Reviewer
Hao Ye, Manuscript Reviewer
When I was asked to review the code for the pavo 2.0 manuscript1, I had an initial moment of panic – I had no experience doing formal code review. Luckily, I knew that rOpenSci had a set of reviewing guidelines, and that a few MEE Applications papers had used them. The same guidelines are also used by the Journal of Open Source Software (JOSS). Although this submission wasn’t flagged for rOpenSci review, I didn’t see a conflict with using their guidelines for my task.
The checklist helped me to organise my review. I started with the basic package review template, and then focused on a detailed look at the primary vignette (which is where I expect most users start). The rOpenSci guidelines encourage the use of some automated tools, like goodpractice to facilitate reviewing. The hardest part was providing suggestions to address what the goodpractice::gp() function flagged as complex or redundant code. The remainder of the review went pretty smoothly. I’m a fan of task checklists, so I’m glad that the authors found my comments useful. Hopefully the changes will help with the future maintenance of the package.
🔗

 Thomas White and Hugo Gruson, Manuscript Authors
Thomas White and Hugo Gruson, Manuscript Authors
We were immediately struck by the rigor and thoughtfulness of the reviews and pleasantly surprised to see reference to rOpenSci in Hao’s [anonymous] review. It was clear that Hao and two other reviewers had invested significant time in examining not only the manuscript and documentation, but the codebase itself. An uncommon, but welcome experience.
Our package was singularly improved as a result, both for end-users and ourselves. Many of the suggestions that we implemented – such as comprehensive test coverage, explicit styling, greater code safety, executable examples, and contributor guidelines – will persist and guide the development of this (and related) packages into the future.
We know that software is challenging to review since the overlap of field-specific expertise between developers and biologists is relatively limited. This is where the value of rOpenSci’s work in developing tractable standards for reviewers and developers really comes into focus, as well as the willingness of journals such as MEE to encourage their use. We’re just grateful for the experience and would be thrilled to see the practice expand in scope and reach where possible.
🔗
 Chris Grieves, Assistant Editor, Methods in Ecology and Evolution
Chris Grieves, Assistant Editor, Methods in Ecology and Evolution
Since the early days of the journal, code and software papers (or Applications articles as we call them) have been really important to MEE. In our Policy on Publishing Code we highlight our commitment to ensuring the quality of code through the peer review process.
We’ve got a team of dedicated Applications Editors who handle code manuscripts and they do a great job of balancing their comments on the manuscript and the code that goes along with it. Resources like the rOpenSci package review guidelines can really help to take the pressure off these Editors, and they give reviewers confidence to comment on the code. It’s great to have the chance to promote them here and we hope that this post will encourage more people to check them out.
We also partner directly with rOpenSci for software peer review. If you have an R package that meets the aims and scope of both MEE and rOpenSci, you can opt for a joint review in which the R package is reviewed by rOpenSci, followed by fast-tracked review of the manuscript by MEE. Manuscripts published through this process are recognized via a mark on both HTML and PDF versions of their paper. We’ve had two articles published to date as a result of this partnership2 3.
🔗 Reflections
Having a manuscript reviewed can often feel like a quite mysterious process. Your work disappears into a black box and comes out with a load of anonymous suggestions for how to improve it. At rOpenSci and Methods in Ecology and Evolution, we want to help open up that black box. Thanks to Hugo’s tweet of gratitude, and the goodwill of the editors, reviewers and authors of the pavo 2.0 paper, this post provides a glimpse of what is possible. Will you give it a try next time?
Maia, R., Gruson, H., Endler, J. A., & White, T. E. (2019). pavo 2: new tools for the spectral and spatial analysis of colour in R. Methods in Ecology and Evolution. https://doi.org/10.1111/2041-210X.13174 ↩︎
Sciaini, M., Fritsch, M., Scherer, C., & Simpkins, C. E. (2018). NLMR and landscapetools: An integrated environment for simulating and modifying neutral landscape models in R. Methods in Ecology and Evolution, 9(11), 2240-2248. https://doi.org/10.1111/2041-210X.13076 ↩︎
Zizka, A., Silvestro, D., Andermann, T., Azevedo, J., Duarte Ritter, C., Edler, D., … & Svantesson, S. CoordinateCleaner: Standardized cleaning of occurrence records from biological collection databases. Methods in Ecology and Evolution. https://doi.org/10.1111/2041-210X.13152 ↩︎

