Tuesday, April 21, 2020 From rOpenSci (https://ropensci.org/blog/2020/04/21/rclean/). Except where otherwise noted, content on this site is licensed under the CC-BY license.
Leave the code cleaner than you found it.
– R.C. Martin in Clean Code
The R language has become very popular among scientists and analysts because it enables the rapid development of software and empowers scientific investigation. However, regardless of the language used, data analysis is usually complicated. Because of various project complexities and time constraints, analytical software often reflects these challenges. “What did I measure? What analyses are relevant to the study? Do I need to transform the data? What’s the function for the analysis I want to run?” Although many researchers see the value in learning to write software, the time investment for learning a programming language alone is still exceedingly high for many, let alone also learning software best practices. The downside to the rapid spread of data science is that learning to create good software takes a back-seat to just writing code that will get the job “done” leading to issues with transparency and software that is highly unstable (i.e. buggy and not reproducible).
The goal of the R package Rclean is to provide a automated tool to help scientists more easily write better code. Specifically, Rclean has been designed to facilitate the isolation of the code needed to produce one or more results, because more often then not, when someone is writing an R script, the ultimate goal is analytical results for inference, such as a set of statistical analyses and associated figures and tables. As the investigative process is inherently iterative, this set of results is nearly always a subset of a much larger set of possible ways to explore a dataset. There are many statistical tests and visualizations and other representations that can be employed in a myriad of ways depending on how the data are processed. This commonly leads to lengthy, complicated scripts from which researchers manually subset results, but which are likely never to be refactored because of the difficulty in disentangling the code.
The Rclean package uses a technique based on data provenance and network algorithms to isolate code for a desired result automatically. The intent is to ease refactoring for scientists that use R but do not have formal training in software design and specifically with the “art” of creating clean code, which in part is the development of an intuitive sense of how code is and/or should be organized. However, manually culling code is tedious and potentially leads to errors regardless of the level of expertise a coder might have; therefore, we see Rclean as a useful tool for programming in R at any level of ability. Here, we’ll cover details on the implementation and design of the package, a general example of how it can be used and thoughts on its future development.
🔗 How to use Rclean to write cleaner code
🔗 Installation
Through the helpful feedback from the rOpenSci community, the package has recently passed software review and a supporting article was recently published in the Journal of Open Source Software 1, in which you can find more details about the package. The package is hosted through the rOpenSci organization on GitHub, and the package can be installed using the devtools package 2 directly from the repository (https://github.com/ROpenSci/Rclean).
library(devtools)
install_github("ROpenSci/Rclean")
If you do not already have RGraphviz, you will need to install it using the following code before installing Rclean:
if (!requireNamespace("BiocManager", quietly = TRUE))
install.packages("BiocManager")
BiocManager::install("Rgraphviz")
🔗 Isolating code for a set of results
Analytical scripts that have not been refactored are often both long and complicated. However, a script doesn’t need to be long to be complicated. The following example script presents some challenges such that even though it’s not a long script, picking through it to get a result would likely prove to be frustrating.
library(stats)
x <- 1:100
x <- log(x)
x <- x * 2
x <- lapply(x, rep, times = 4)
### This is a note that I made for myself.
### Next time, make sure to use a different analysis.
### Also, check with someone about how to run some other analysis.
x <- do.call(cbind, x)
### Now I'm going to create a different variable.
### This is the best variable the world has ever seen.
x2 <- sample(10:1000, 100)
x2 <- lapply(x2, rnorm)
### Wait, now I had another thought about x that I want to work through.
x <- x * 2
colnames(x) <- paste0("X", seq_len(ncol(x)))
rownames(x) <- LETTERS[seq_len(nrow(x))]
x <- t(x)
x[, "A"] <- sqrt(x[, "A"])
for (i in seq_along(colnames(x))) {
set.seed(17)
x[, i] <- x[, i] + runif(length(x[, i]), -1, 1)
}
### Ok. Now I can get back to x2.
### Now I just need to check out a bunch of stuff with it.
lapply(x2, length)[1]
max(unlist(lapply(x2, length)))
range(unlist(lapply(x2, length)))
head(x2[[1]])
tail(x2[[1]])
## Now, based on that stuff, I need to subset x2.
x2 <- lapply(x2, function(x) x[1:10])
## And turn it into a matrix.
x2 <- do.call(rbind, x2)
## Now, based on x2, I need to create x3.
x3 <- x2[, 1:2]
x3 <- apply(x3, 2, round, digits = 3)
## Oh wait! Another thought about x.
x[, 1] <- x[, 1] * 2 + 10
x[, 2] <- x[, 1] + x[, 2]
x[, "A"] <- x[, "A"] * 2
## Now, I want to run an analysis on two variables in x2 and x3.
fit.23 <- lm(x2 ~ x3, data = data.frame(x2[, 1], x3[, 1]))
summary(fit.23)
## And while I'm at it, I should do an analysis on x.
x <- data.frame(x)
fit.xx <- lm(A~B, data = x)
summary(fit.xx)
shapiro.test(residuals(fit.xx))
## Ah, it looks like I should probably transform A.
## Let's try that.
fit_sqrt_A <- lm(I(sqrt(A))~B, data = x)
summary(fit_sqrt_A)
shapiro.test(residuals(fit_sqrt_A))
## Looks good!
## After that. I came back and ran another analysis with
## x2 and a new variable.
z <- c(rep("A", nrow(x2) / 2), rep("B", nrow(x2) / 2))
fit_anova <- aov(x2 ~ z, data = data.frame(x2 = x2[, 1], z))
summary(fit_anova)
So, let’s say we’ve come to our script wanting to extract the code to
produce one of the results fit_sqrt_A, which is an analysis that is
the fitted model object for an analysis. We might want to double check
the results, and we also might need to use the code again for another
purpose, such as creating a plot of the patterns supported by the
test. Manually tracing through our code for all the variables used in
the test and finding all of the code used to prepare them
for the analysis would be difficult, especially given the
fact that we have used “x” as a prefix for multiple unrelated objects
in the script. However, Rclean can
do this easily via the clean() function.
library(Rclean)
script <- system.file("example",
"long_script.R",
package = "Rclean")
clean(script, "fit_sqrt_A")
x <- 1:100
x <- log(x)
x <- x * 2
x <- lapply(x, rep, times = 4)
x <- do.call(cbind, x)
x <- x * 2
colnames(x) <- paste0("X", seq_len(ncol(x)))
rownames(x) <- LETTERS[seq_len(nrow(x))]
x <- t(x)
x[, "A"] <- sqrt(x[, "A"])
for (i in seq_along(colnames(x))) {
set.seed(17)
x[, i] <- x[, i] + runif(length(x[, i]), -1, 1)
}
x[, 1] <- x[, 1] * 2 + 10
x[, 2] <- x[, 1] + x[, 2]
x[, "A"] <- x[, "A"] * 2
x <- data.frame(x)
fit_sqrt_A <- lm(I(sqrt(A)) ~ B, data = x)
The output is the code that Rclean has picked out from the tangled bits of code, which in this case is an example script included with the package. Here’s a view of this isolated code highlighted in the original script.
library(stats)
x <- 1:100
x <- log(x)
x <- x * 2
x <- lapply(x, rep, times = 4)
### This is a note that I made for myself.
### Next time, make sure to use a different analysis.
### Also, check with someone about how to run some other analysis.
x <- do.call(cbind, x)
### Now I'm going to create a different variable.
### This is the best variable the world has ever seen.
x2 <- sample(10:1000, 100)
x2 <- lapply(x2, rnorm)
### Wait, now I had another thought about x that I want to work through.
x <- x * 2
colnames(x) <- paste0("X", seq_len(ncol(x)))
rownames(x) <- LETTERS[seq_len(nrow(x))]
x <- t(x)
x[, "A"] <- sqrt(x[, "A"])
for (i in seq_along(colnames(x))) {
set.seed(17)
x[, i] <- x[, i] + runif(length(x[, i]), -1, 1)
}
### Ok. Now I can get back to x2.
### Now I just need to check out a bunch of stuff with it.
lapply(x2, length)[1]
max(unlist(lapply(x2, length)))
range(unlist(lapply(x2, length)))
head(x2[[1]])
tail(x2[[1]])
## Now, based on that stuff, I need to subset x2.
x2 <- lapply(x2, function(x) x[1:10])
## And turn it into a matrix.
x2 <- do.call(rbind, x2)
## Now, based on x2, I need to create x3.
x3 <- x2[, 1:2]
x3 <- apply(x3, 2, round, digits = 3)
## Oh wait! Another thought about x.
x[, 1] <- x[, 1] * 2 + 10
x[, 2] <- x[, 1] + x[, 2]
x[, "A"] <- x[, "A"] * 2
## Now, I want to run an analysis on two variables in x2 and x3.
fit.23 <- lm(x2 ~ x3, data = data.frame(x2[, 1], x3[, 1]))
summary(fit.23)
## And while I'm at it, I should do an analysis on x.
x <- data.frame(x)
fit.xx <- lm(A~B, data = x)
summary(fit.xx)
shapiro.test(residuals(fit.xx))
## Ah, it looks like I should probably transform A.
## Let's try that.
fit_sqrt_A <- lm(I(sqrt(A))~B, data = x)
summary(fit_sqrt_A)
shapiro.test(residuals(fit_sqrt_A))
## Looks good!
## After that. I came back and ran another analysis with
## x2 and a new variable.
z <- c(rep("A", nrow(x2) / 2), rep("B", nrow(x2) / 2))
fit_anova <- aov(x2 ~ z, data = data.frame(x2 = x2[, 1], z))
summary(fit_anova)
The isolated code can now be visually inspected to adapt the original
code or ported to a new, refactored script using keep().
fitSA <- clean(script, "fit_sqrt_A")
keep(fitSA)
This will pass the code to the clipboard for pasting into another document. To write directly to a new file, a file path can be specified.
fitSA <- clean(script, "fit_sqrt_A")
keep(fitSA, file = "fit_SA.R")
To explore more possible variables to extract, the get_vars() function
can be used to produce a list of the variables (aka. objects) that are
created in the script.
get_vars(script)
[1] "x" "x2" "i" "x3" "fit.23"
[6] "fit.xx" "fit_sqrt_A" "z" "fit_anova"
Especially when the code for different variables are entangled, it can
be useful to visual the code in order to devise an approach to
cleaning. The code_graph() function can also give us a visual of the
code and the objects that they produce.
code_graph(script)
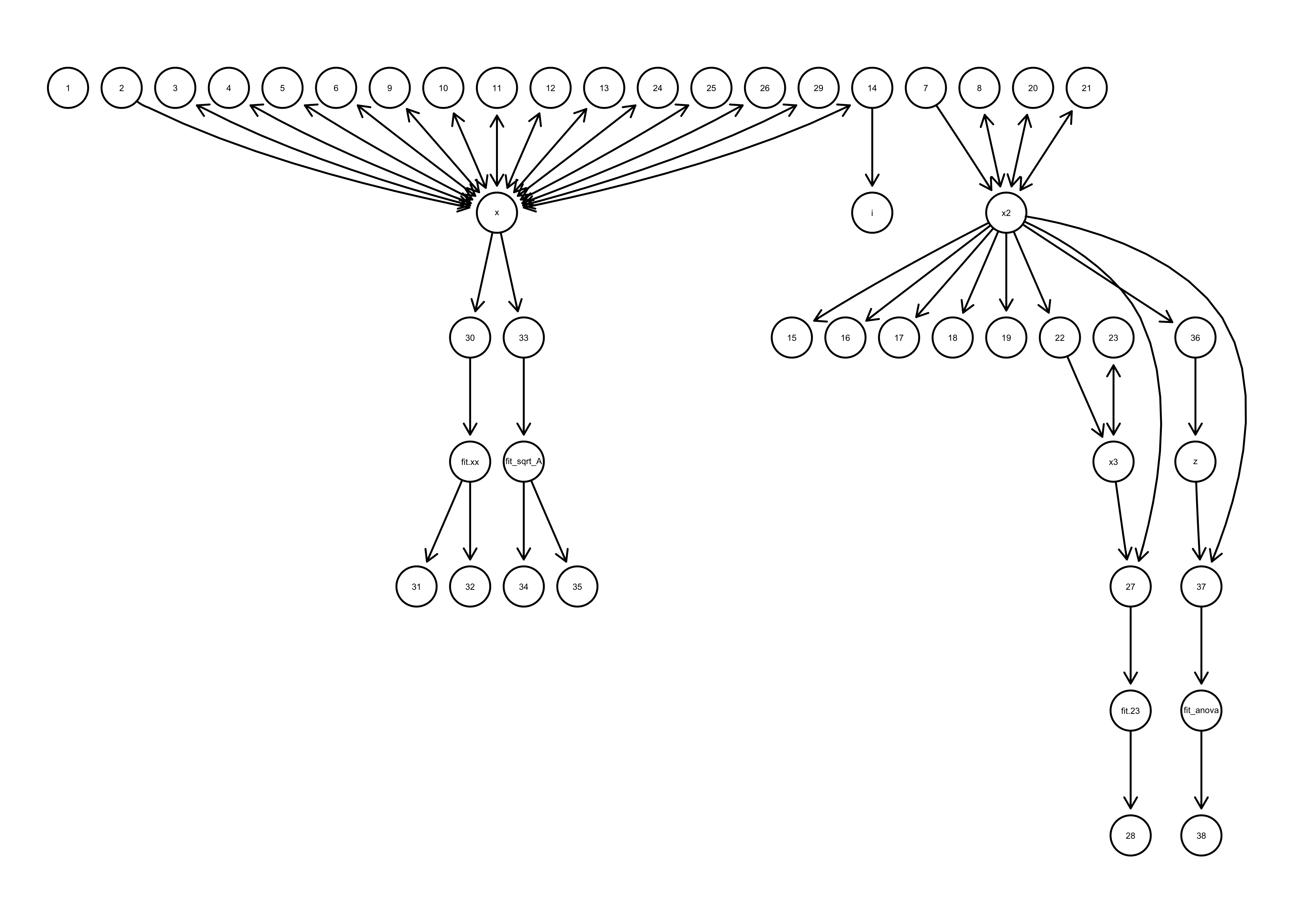
Figure 1 code_graph() example: Example of the plot produced by the code_graph function showing which functions produce which variables and which variables are used as inputs to other functions.
After examining the output from get_vars() and code_graph(), it is
possible that more than one object needs to be isolated. This can be
done by adding additional objects to the list of vars.
clean(script, vars = c("fit_sqrt_A", "fit_anova"))
x <- 1:100
x <- log(x)
x <- x * 2
x <- lapply(x, rep, times = 4)
x <- do.call(cbind, x)
x2 <- sample(10:1000, 100)
x2 <- lapply(x2, rnorm)
x <- x * 2
colnames(x) <- paste0("X", seq_len(ncol(x)))
rownames(x) <- LETTERS[seq_len(nrow(x))]
x <- t(x)
x[, "A"] <- sqrt(x[, "A"])
for (i in seq_along(colnames(x))) {
set.seed(17)
x[, i] <- x[, i] + runif(length(x[, i]), -1, 1)
}
x2 <- lapply(x2, function(x) x[1:10])
x2 <- do.call(rbind, x2)
x[, 1] <- x[, 1] * 2 + 10
x[, 2] <- x[, 1] + x[, 2]
x[, "A"] <- x[, "A"] * 2
x <- data.frame(x)
fit_sqrt_A <- lm(I(sqrt(A)) ~ B, data = x)
z <- c(rep("A", nrow(x2) / 2), rep("B", nrow(x2) / 2))
fit_anova <- aov(x2 ~ z, data = data.frame(x2 = x2[, 1], z))
Currently, libraries can not be isolated directly during the cleaning
process. So, the get_libs() function provides a way to detect the
libraries for a given script. We just need to supply a file path and
get_libs() will return the libraries that are called by that script.
get_libs(script)
[1] "stats"
🔗 The provenance engine under the hood
The clean() function provides an effective way to remove code that is
unwanted; however, many researchers are wary about doing this exact
thing for at least a few reasons. Perhaps the top reason is that the
main goal of an analysis is the results and taking time to craft
transparent, dependable software is not the priority. As such, taking
time to go back through a script and remove code is time
wasted. Relatedly, for most researchers the best way to keep track of
the various analyses that they have explored is to keep them in the
script, as they do not use a rigorous version control system but
instead rely on file backups and informal versioning. Although we
can’t give researchers more hours in the day, providing an easier and
more reliable means to remove unused code will lower the barrier to
creating better, cleaner code. Combined with the increasing use of
version control systems and digital notebooks, the practice of
“saving” analytical ideas in a script will become less common and code
quality will increase.
The process that Rclean uses
relies on the generation of data provenance. The term provenance
means information about the origins of some object. Data provenance is
a formal representation of the execution of a computational process, to rigorously determine the the
unique computational pathway from inputs to results 3. To
avoid confusion, note that “data” in this context is used in a broad
sense to include all of the information generated during computation,
not just the data that are collected in a research project that are
used as input to an analysis. Having the formalized, mathematically
rigorous representation that data provenance provides guarantees that
analyses conducted by Rclean
are theoretically sound. Most importantly, because the relationships
defined by the provenance can be represented as a graph, it is
possible to apply network search algorithms to determine the minimum
and sufficient code needed to generate the chosen result in the
clean() function.
There are multiple approaches to collecting data provenance, but
Rclean uses “prospective”
provenance, which analyzes code and uses language-specific information
to predict the relationship among processes and data
objects. Rclean relies on an
R package called CodeDepends to gather the prospective provenance for
each script. For more information on the mechanics of the CodeDepends
package, see 4. To get an idea of what data provenance
is, we can use the code_graph() function to get a graphical representation
of the prospective provenance generated for
Rclean.
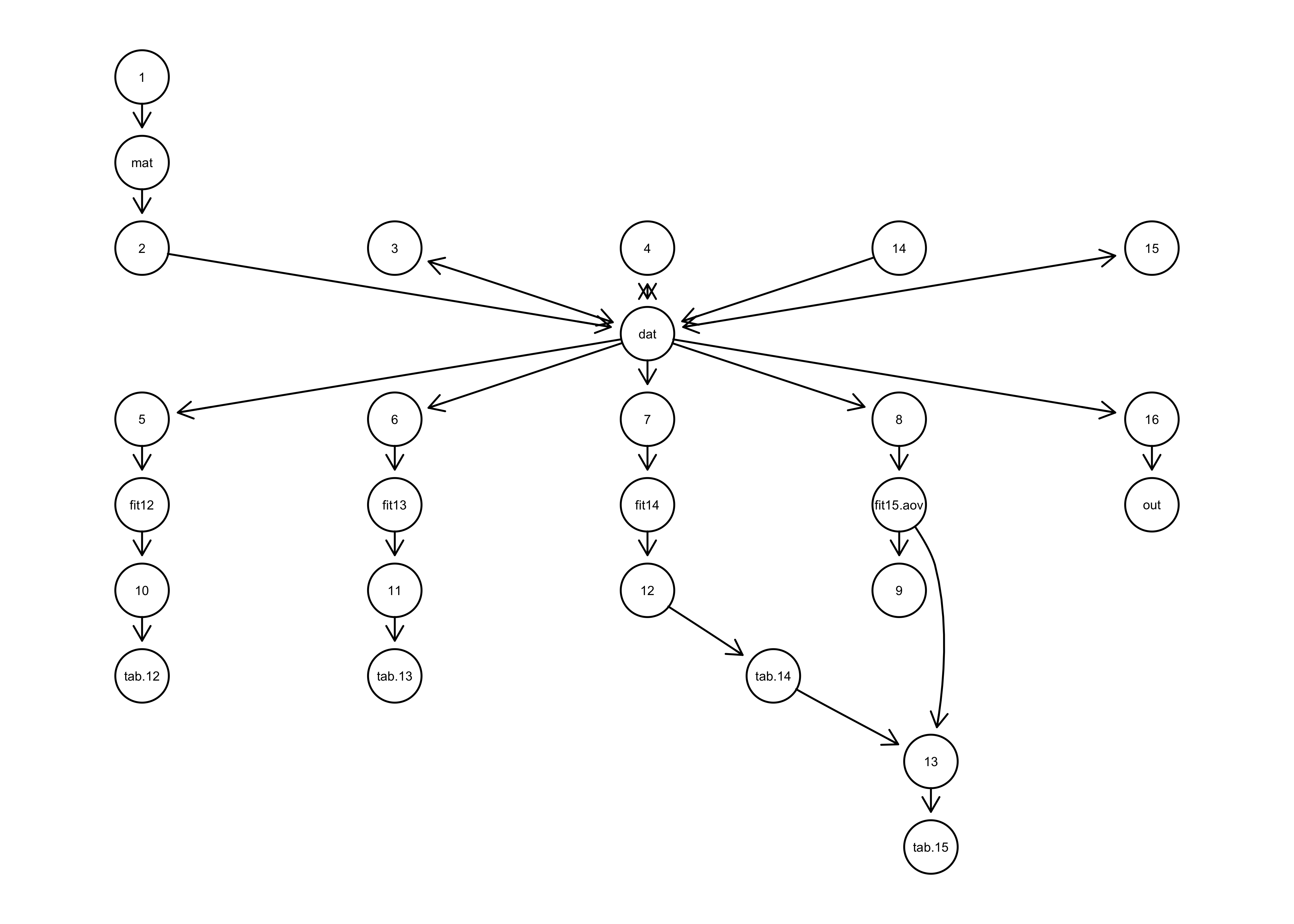
Figure 2 provenance graph: Network diagram of the prospective data provenance generated for an example script. Arrows indicate which functions (numbers) produced (outgoing arrow) or used (incoming arrow) which objects (names).
All of this work with the provenance is to get the network
representation of relationships among functions and objects. The
provenance network is very powerful because we can now apply
algorithms to analyze the R script with respect to our results. This is
what empowers the clean() function, which takes the provenance and
applies a network search algorithm to determine the pathways leading
from inputs to outputs. In the process any objects or functions
that do not fall along that pathway are by definition not necessary to
produce the desired set of results and can therefore be removed. As
demonstrated in the example, this property of the provenance network
is what facilitates the robust isolation of the minimal code necessary
to generate the output we want.
One important topic to discuss is that Rclean does not keep comments present in code. This is the result of a limitation of the data provenance collection, which currently does not assign them a relationship in the provenance network. This is a general issue with detecting the relationships between comments and code. For example, comments at the end of lines are typically relevant to the line they are on but this is not a linguistic requirement. Also, comments occupying their own lines usually refer to the following lines but this is also not necessarily the case either. In fact comments can refer to any or none of the code relative to their position in the script, the latter commonly being the case when code is removed from a script but comments referring to it have not. The inferred and explicit meanings of comments are a cultural and not linguistic.
That being said, although Rclean
cannot operate automatically on comments, comments in the original
code remain untouched and can be used to inform the reduced
code. Also, as the clean() function is oriented toward isolating
code based on a specific result, the resulting code tends to naturally
support the generation of new comments that are higher level
(e.g. “The following produces a plot of the mean response of each
treatment group.”), and lower level comments are not necessary because
the code is simpler and clearer. This process of commenting is an
important part of writing better code. Lastly, although comments can
serve an important role in coding, it is worth reflecting on the
statement in R.C. Martin’s book Clean Code: A Handbook of Agile
Software Craftsmanship where he writes that, “Comments do not
compensate for bad code.”
🔗 Concluding remarks and future work
Rclean provides a simple, easy to use tool for scientists who would like help refactoring code. Using Rclean, the code necessary to produce a specified result (e.g., an object stored in memory or a table or figure written to disk) can be easily and reliably isolated even when tangled with code for other results. Tools, such as this, that make it easier to produce transparent, accessible code will be an important aid for improving scientific reproducibility 5.
Although the current implementation of Rclean for minimizing code is useful on its own, we see promise in connecting it with other reproducibility tools. One example is the reprex package, which provides a simple API for sharing reproducible examples 6. Rclean could provide a reliable way to extract parts of a larger script that would be piped to a simplified reproducible example. Another possibility is to help transition scripts to functions, packages and workflows refactoring via toolboxes like drake 7. Since Rclean can isolate the code from inputs to one or more outputs, it could be used to extract all of the components needed to write one or more functions that would be a part of a package or workflow, as is the goal of drake.
In the future, it would also be useful to extend the existing framework to support other provenance methods. One possibility is retrospective provenance, which tracks a computational process as it is executing. Through this active, concurrent monitoring, retrospective provenance can gather information that static prospective provenance can’t. Greater details of the computational process would enable other features that could address some challenges, such as libraries that are actually used by the code, processing comments (as discussed above), parsing control statements and replicating random processes. Using retrospective provenance comes at a cost, however. In order to gather it, the script needs to be executed. When scripts are computationally intensive or contain bugs that stop execution retrospective provenance can not be obtained for part or all of the code. Although such costs may present challenges, combining prospective and retrospective provenance methods could provide a powerful and flexible solution. Some work has already been done in the direction of implementing retrospective provenance for code cleaning in R (see http://end-to-end-provenance.github.io); however, there doesn’t appear to be a tool that synthesizes these two approaches to provenance.
We hope that Rclean makes writing scientific software easier for the R community. The package has already been significantly improved via the rOpenSci review process, thanks to the efforts of editor Anna Krystalli and reviewers, Will Landau and Clemens Schmid. We look forward to the future progress of the package and other “code cleaning” tools. As an open-source project, we would like to encourage feedback and help with extending the package. We invite people to use the package and get involved by reporting bugs and suggesting or (hopefully) contributing features. For more information please visit the project page on GitHub.
🔗 Acknowledgements
Thanks to Steffi Lazerte, Stefanie
Butland and Maëlle
Salmon for editorial work and technical
help. Special word of thanks to Maëlle
Salmon for the addition of the code
highlighting figure in the clean() example.
Lau M, Pasquier TFJ, Seltzer M (2020). “Rclean: A Tool for Writing Cleaner, More Transparent Code.” Journal of Open Source Software, 5(46), 1312. https://doi.org/10.21105/joss.01312 , https://doi.org/10.21105/joss.01312. ↩︎
Wickham H, Hester J, Chang W (2019). devtools: Tools to Make Developing R Packages Easier. https://CRAN.R-project.org/package=devtools. ↩︎
Carata L, Akoush S, Balakrishnan N, Bytheway T, Sohan R, Seltzer M, Hopper A (2014). “A Primer on Provenance.” Queue, 12(3), 10-23. ISSN 15427730, http://dl.acm.org/citation.cfm?doid=2602649.2602651 , https://doi.org/10.1145/2602649.2602651. ↩︎
Temple Lang D, Peng R, Nolan D, Becker G (2020). CodeDepends: Analysis of R Code for Reproducible Research and Code Comprehension. https://github.com/duncantl/CodeDepends. ↩︎
Pasquier T, Lau MK, Trisovic A, Boose ER, Couturier B, Crosas M, Ellison AM, Gibson V, Jones CR, Seltzer M (2017). “If these data could talk.” Scientific Data, 4, 170114. ISSN 2052-4463, http://www.nature.com/articles/sdata2017114 , https://doi.org/10.1038/sdata.2017.114. ↩︎
Bryan J, Hester J, Robinson D, Wickham H (2019). reprex: Prepare Reproducible Example Code via the Clipboard. https://CRAN.R-project.org/package=reprex. ↩︎
Landau WM (2020). drake: A Pipeline Toolkit for Reproducible Computation at Scale. https://CRAN.R-project.org/package=drake. ↩︎

