Tuesday, October 20, 2020 From rOpenSci (https://ropensci.org/blog/2020/10/20/searching-medrxivr-and-biorxiv-preprint-data/). Except where otherwise noted, content on this site is licensed under the CC-BY license.
Background & motivation
medRxiv, as the preprint repository for papers in the medical, clinical, and related health sciences,1 has become a central source of new studies related to the COVID-19 pandemic. As a result, more and more researchers have begun to include medRxiv in the list of bibliographic databases they search as part of systematic reviews, a type of study that aims to find and bring together all available evidence on a topic in order to provide a comprehensive answer to a research question.
However, the native search functionality available on the medRxiv website is not suitable for use in systematic reviews due to a number of limitations. This prompted the development of medrxivr, an R package which provides access to, and tools for searching, medRxiv and bioRxiv preprint metadata. This post will detail the limitations of the native medRxiv search and summarize the key functionality of medrxivr via two key use cases. Note that while medrxivr allows users to access and search medrxiv and bioRxiv preprint metadata as both repositories are run by the Cold Spring Harbor Laboratory and so have a similar API endpoint/native website search functionality, all illustrating examples in this post are taken from medRxiv.
🔗 Limitations of medRxiv website search functionality
🔗 Confusing information about Boolean operators
A key limitation of the native search on the medRxiv website comes from the use of Boolean operators in search strings. Boolean operators (AND, OR, NOT) are used by librarians to combine individual search terms into specific complex search strings. For example, “(dementia OR alzheimer)” would be expected to return any record containing either “dementia” or “alzheimer”. Combining this with a second topic via AND, e.g. “(dementia OR alzheimer) AND (lipid)”, should return any record containing either “dementia” or “alzheimer” once it also contains the term “lipid”.
One of the key assumptions of Boolean operators is that the order of the terms does not matter, i.e. that “(dementia OR alzheimer) AND lipids” is equivalent to “(alzheimer OR dementia) AND lipids”. However, running these searches on the 13th Oct 2020, the first gives 133 results and the second gives 126. It is not clear what is happening behind the scenes, but there is potentially some weighting of terms based on proximity. This means that key records may be missed based on the arbitrary ordering of the search terms.
🔗 Lack of reproducbility
One of the key tenets of systematic reviews is that literature searches are transparent and reproducible - that is, that a certain search strategy (e.g. “(dementia OR alzheimer) AND lipids”) will provide the same number of results each time it is performed. However, anecdotal evidence from information specialists and medical librarians suggests that medRxiv’s web search returns inconsistent citations over time.
🔗 No batch export
Any systematic reviewer will tell you that for a database to be useful as an information source, the one thing it needs to have is an option to export the results of a given search en masse. At present, this is not something the medRxiv website supports.
This means that records returned by a search must be exported by hand in small batches, an error-prone and time-consuming (not to mention anger-inducing!) method of extracting citations from a bibliographic database.
🔗 Use cases
In light of these limitations, below are two key use cases for the medrxivr package.
🔗 Use case #1 – Get preprint metadata in R
The primary aim of medrxivr is to make it as easy as possible to get cleaned preprint metadata into R. This enables a range of exploratory analysis to be readily performed. For example, to explore the distribution of topic categories for those preprints posted to medRxiv in the first week of 2020, the following code can be used:
# Load the packages
library(medrxivr)
library(dplyr)
library(ggplot2)
# Import the data from medRxiv API endpoint
mx_data <- mx_api_content(from_date = "2020-01-01", to_date = "2020-01-07")
# Create the graph
mx_data %>%
group_by(category) %>%
summarise(N = n(), .groups = "keep") %>%
arrange(desc(N)) %>%
ggplot(aes(y = reorder(category,N), x = N)) +
geom_col(colour = "black") +
ylab("Category") +
xlab("Number of preprints, 1st week of January 2020") +
scale_x_continuous(expand = c(0, 0), limits = c(0,10)) +
theme_classic()
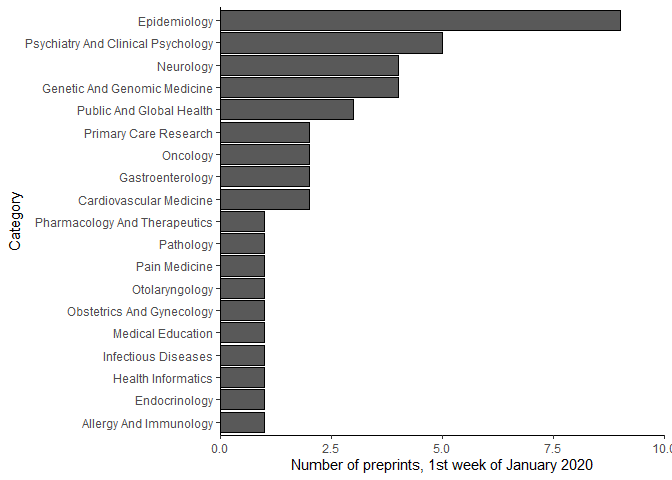
🔗 Use case #2 – Reproducible searching and exporting
While being able to easily import medRxiv data is useful, as a systematic reviewer, my main interest is in what medrxivr allows you to do once you have imported a local copy of the database. As mentioned in the section above, the native medRxiv website search interface is neither reproducible nor transparent, and while the API (accessed via mx_api_content()) is great for accessing medRxiv metadata, it does not offer any search functionality. The second core aspect of medrxivr’s functionality is designed to address these limitations, allowing information specialists to build complex searches and apply them to a local copy of the medRxiv database, all while documenting their search strategy in a transparent manner. An example is given below, where a researcher is looking for records that contain both dementia-related and lipid-related terms:
# Use the maintained snapshot to quickly load today's copy of the medRxiv database
# Note - this `preprint_data` object could be saved as a CSV in your project
# repository to aid reproducibility
preprint_data <- mx_snapshot()
## Using medRxiv snapshot - 2020-10-19 00:32
# Build your search
topic1 <- c("dementia","vascular","alzheimer's") # Combined with Boolean OR
topic2 <- c("lipids","statins","cholesterol") # Combined with Boolean OR
myquery <- list(topic1, topic2) # Combined with Boolean AND
# Run your search
results <- mx_search(data = preprint_data,
query = myquery)
## Found 48 record(s) matching your search.
# Let's have a look at the returned records
results
## # A tibble: 48 x 14
## ID title abstract authors date category doi version author_correspo~ author_correspo~
## <dbl> <chr> <chr> <chr> <date> <chr> <chr> <dbl> <chr> <chr>
## 1 184 Exce~ "Object~ De Lim~ 2019-07-25 Cardiov~ 10.1~ 1 Andrei C Sposito "Laboratory of ~
## 2 281 Peri~ "Introd~ Newton~ 2019-08-14 Addicti~ 10.1~ 1 Dwight F Newton "University of ~
## 3 284 A co~ "Object~ Moriar~ 2019-08-15 Cardiov~ 10.1~ 1 Mark H Ebell "University of ~
## 4 341 Apol~ "Backgr~ Richar~ 2019-08-29 Cardiov~ 10.1~ 1 Tom G Richardson "University of ~
## 5 307 Comp~ "Owing ~ Batty,~ 2019-08-17 Epidemi~ 10.1~ 1 George David Ba~ "University Col~
## 6 473 Self~ "Backgr~ Grover~ 2019-09-23 Cardiov~ 10.1~ 1 Abhinav Grover "University of ~
## 7 643 Tren~ "Object~ Curtis~ 2019-10-18 Cardiov~ 10.1~ 1 Ben Goldacre "University of ~
## 8 978 Does~ "OBJECT~ Harber~ 2019-11-29 Endocri~ 10.1~ 1 Lisa Harber-Asc~ "King\\'s Colle~
## 9 1085 Asse~ "Backgr~ Finner~ 2019-12-14 Cardiov~ 10.1~ 1 Pradeep Nataraj~ "Massachusetts ~
## 10 1333 Idea~ "BACKGR~ Mckenz~ 2020-01-16 Epidemi~ 10.1~ 1 Trevor S Fergus~ "Caribbean Inst~
## # ... with 38 more rows, and 4 more variables: link_page <chr>, link_pdf <chr>, license <chr>,
## # published <chr>
Once you have run your search, exporting the results to a .BIB file for import into a reference manager, such as Zotero or Mendeley, is as simple as passing the results object to the mx_export() function.
Similarly, as screening the full text PDFs of records returned by a search against eligibility criteria forms a key part of a systematic review, a second helper function downloads the PDF for each record returned by mx_search(). Again, it is simply a case of passing the results object to the mx_download() function.
🔗 Conclusion
medRxiv is a fantastic resource and has been a key source of information related to the COVID-19 pandemic. However, some key issues with the website mean that its native search functionality precludes its use in systematic reviews. The medrxivr R package seeks to address these limitations by providing a user-friendly way to import and systematically search medRxiv and bioRxiv records in R. Full documentation of the package functionality, particularly around the implementation of complex search strategies using syntax such as wildcards and the NEAR operator, which is used to find co-located terms (e.g. “cholesterol NEAR2 test” find records where “cholesterol” and “test” are separated by two or less words), is available from the medrxivr website. Finally, I wanted to note that during development, medrxivr benefited immensely from an rOpenSci peer review by Tuija Sonkkila and Najko Jahn.
Rawlinson, C., & Bloom, T. (2019). New preprint server for medical research. BMJ, 365. https://doi.org/doi:10.1136/bmj.l2301 ↩︎

