Friday, April 22, 2022 From rOpenSci (https://ropensci.org/blog/2022/04/22/ropensci-news-digest-april-2022/). Except where otherwise noted, content on this site is licensed under the CC-BY license.
Dear rOpenSci friends, it’s time for our monthly news roundup!
You can read this post on our blog. Now let’s dive into the activity at and around rOpenSci!
🔗 rOpenSci HQ
🔗 pkgcheck reports now include dependencies
Our automated checking system for packages submitted for peer review now has a new section, “Package Dependencies”.
This is intended to help editors and reviewers understand how package dependencies (that is, packages listed in Imports, Depends, or Suggests DESCRIPTION fields) are actually used by a package.
The section summarises total numbers of function calls made to each package, followed by collapsible “details” sections containing numbers of calls made to the individual functions of each packages.
Dependencies include base R and all recommended packages, ensuring that this section provides a comprehensive overview of how each package submitted for peer review uses and depends upon the entire R ecosystem.
Want to try it for yourself?
Install pkgcheck and then run pkgcheck::pkgcheck (<package-source-directory>).
🔗 R-universe now features package individual pages
Packages on r-universe.dev now have beautiful individual landing pages, which give an overview of the current content and metadata for that package. This includes the package description, author information, articles, download links, commits, tags, contributors, reference manual, cff citation, etc. We are also working on adding more information about usage of the package, such as citations and reverse dependencies.
You can navigate to a package homepage directly from search results or via the dashboard from its universe. It is also possible to link directly to the package homepage, for example: https://ropensci.r-universe.dev/ui#package:cffr
🔗 Next coworking sessions
Join us for social coworking & office hours monthly on 1st Tuesdays! Hosted by Steffi LaZerte and various community hosts. Everyone welcome. No RSVP needed. Consult our Events page to find your local time and how to join.
Our next sessions are:
Tuesday, 03 May 2022 9 AM North American Pacific / 16:00 UTC “Why be a reviewer of R packages?” Hosted by community host François Michonneau and Steffi LaZerte
- Cowork on a project of your choice;
- Prepare an R package for rOpenSci software review, work on reviewer comments, or review a package;
- Or ask François Michonneau what it’s like to review packages!
Tuesday, 07 June 2022 9 AM Australian Western / 1:00 UTC “Tackle something you’ve always wanted to learn” Hosted by community host Nick Tierney and Steffi LaZerte
- Cowork on a project of your choice;
- Take this time to tackle something you’ve always wanted to learn;
- Or talk to Nick Tierney and discuss what it is you want to learn, and get some tips and resources to get yourself started!
Find out about more events.
🔗 Software 📦
🔗 New packages
The following three packages recently became a part of our software suite:
gbifdb, developed by Carl Boettiger: A high performance interface to the Global Biodiversity Information Facility, GBIF. In contrast to rgbif, which can access small subsets of GBIF data through web-based queries to a central server, gbifdb provides enhanced performance for R users performing large-scale analyses on servers and cloud computing providers, providing full support for arbitrary SQL or dplyr operations on the complete GBIF data tables (now over 1 billion records, and over a terabyte in size). gbifdb accesses a copy of the GBIF data in parquet format, which is already readily available in commercial computing clouds such as the Amazon Open Data portal and the Microsoft Planetary Computer, or can be accessed directly without downloading, or downloaded to any server with suitable bandwidth and storage space.
qualR, developed by Mario Gavidia-Calderón together with Daniel Schuch: A package to download information from CETESB QUALAR https://cetesb.sp.gov.br/ar/qualar/ and MonitorAr https://www.rio.rj.gov.br/web/smac/monitorar-rio1 systems. It contains function to download different parameters, a set of criteria pollutants and the most frequent meteorological parameters used in air quality data analysis and air quality model evaluation. It has been reviewed by Beatriz Milz, and Kaue de Sousa.
rsvg, developed by Jeroen Ooms: Renders vector-based svg images into high-quality custom-size bitmap arrays using librsvg2. The resulting bitmap can be written to e.g. png, jpeg or webp format. In addition, the package can convert images directly to various formats such as pdf or postscript. It is available on CRAN.
Discover more packages, read more about Software Peer Review.
🔗 New versions
The following sixteen packages have had an update since the last newsletter: gert (v1.6.0), antiword (v1.3.1), baRcodeR (v0.1.7), cffr (v0.2.2), comtradr (0.3.0), git2rdata (v0.4.0), MODIStsp (v2.0.7), nasapower (v4.0.7), occCite (v0.5.4), pdftools (v3.2.0), rgbif (v3.7.2), rsvg (v2.3.1), tarchetypes (0.6.0), targets (0.12.0), taxa (v0.4.2), and tidyhydat (0.5.5).
🔗 Software Peer Review
There are sixteen recently closed and active submissions and 4 submissions on hold. Issues are at different stages:
Two at ‘6/approved’:
EDIutils, An API Client for the Environmental Data Initiative Repository. Submitted by Colin Smith.
gbifdb, Local Database Interface to GBIF. Submitted by Carl Boettiger.
One at ‘5/awaiting-reviewer(s)-response’:
- ReLTER, An interface to the eLTER for the R statistical programming environment. Submitted by Alessandro Oggioni.
Four at ‘4/review(s)-in-awaiting-changes’:
npi, Access the U.S. National Provider Identifier Registry API. Submitted by Frank Farach.
phruta, Phylogenetic Reconstruction and Time-dating. Submitted by Cristian Román-Palacios.
epair, Grabs data from EPA API, simplifies getting pollutant data. Submitted by Leo Orozco-Mulfinger.
healthdatacsv, Access data in the healthdata.gov catalog. Submitted by iecastro.
Five at ‘3/reviewer(s)-assigned’:
unifir, A Unifying API for Calling Unity from R. Submitted by Michael Mahoney.
canaper, Categorical Analysis of Neo- And Paleo-Endemism. Submitted by Joel Nitta. (Stats).
tidyqpcr, Quantitative PCR Analysis with the Tidyverse. Submitted by Edward Wallace.
tsbox, Class-Agnostic Time Series. Submitted by Christoph Sax. (Stats).
ROriginStamp, Interface to OriginStamp API to Obtain Trusted Time Stamps. Submitted by Rainer M Krug.
One at ‘2/seeking-reviewer(s)’:
- octolog, Better Github Action Logging. Submitted by Jacob Wujciak-Jens.
Three at ‘1/editor-checks’:
hudr, A R interface for accessing HUD (US Department of Housing and Urban Development) APIs. Submitted by Emmet Tam.
bssm, Bayesian Inference of Non-Linear and Non-Gaussian State Space. Submitted by Jouni Helske. (Stats).
rdbhapi, Interface to DBH-API. Submitted by Marija Ninic.
Find out more about Software Peer Review and how to get involved.
🔗 On the blog
🔗 Software Review
- Editorial Challenges and Solutions in Software Peer Review by Maëlle Salmon, and Noam Ross. Challenges encountered by editors, and the measures we’ve taken to try to alleviate said problems.
🔗 Tech Notes
Searching and browsing the R universe by Jeroen Ooms. We made a series of structural improvements in r-universe to make it easier to browse and discover interesting R packages and articles. Most notably, the landing page has been overhauled: you can now search directly for any name or keyword across the entire ecosystem.
rsnps 0.5.0: New ncbi_snp_query() Features by Sina Rüeger, and Julia Gustavsen.
ncbi_snp_query()now returns all reported variant allele frequencies in dbSNP.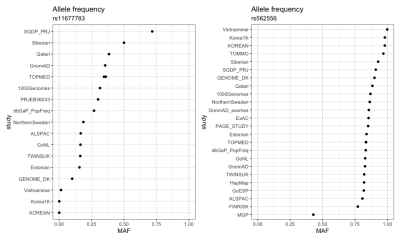
Safeguards and Backups for GitHub Organizations by Maëlle Salmon. Some practical steps we’ve taken to safeguard our GitHub organizations.
🔗 Use cases
Two use cases of our packages and resources have been reported since we sent the last newsletter.
Using vcr for HTTP testing of a package for Kobotoolbox. Reported by Frie.
Estimating leaf temperatures worldwide. Reported by Paul Melloy.
Explore other use cases and report your own!
🔗 Call for maintainers
There are no open calls for new maintainers at this point but you can refer to our contributing guide for finding ways to get involved!
As the maintainer of an rOpenSci package, feel free to contact us on Slack or email [email protected] to get your call for maintainer featured in the next newsletter.
🔗 Package development corner
Some useful tips for R package developers. 👀
🔗 Identifying code duplication
Are you worried a codebase you’ve just inherited doesn’t follow the DRY (don’t repeat yourself) principle? You might find the dupree package by Russ Hyde useful: “The goal of dupree is to identify chunks / blocks of highly duplicated code within a set of R scripts.”.
🔗 Security for package developers
Did you know our development guide includes a short chapter on Package Development Security Best Practices? Starting with the recommendation to enable two-factor authentication (2FA) for your GitHub account (now required for the ropensci GitHub organization). We’ve also recently blogged on Safeguards and Backups for GitHub Organizations.
🔗 S3, S4, R6, R7 (not an April Fools’ joke!)
Have you heard of the R7 package? It is meant to be an object-oriented programming system successor to S3 and S4. It is developped under the umbrella of an R Consortium working group, and a long term goal is to merge it into base R. Explore the current repository and get a taste of the future!
🔗 Last words
Thanks for reading! If you want to get involved with rOpenSci, check out our Contributing Guide that can help direct you to the right place, whether you want to make code contributions, non-code contributions, or contribute in other ways like sharing use cases.
If you haven’t subscribed to our newsletter yet, you can do so via a form. Until it’s time for our next newsletter, you can keep in touch with us via our website and Twitter account.

