Monday, February 27, 2023 From rOpenSci (https://ropensci.org/blog/2023/02/27/runiverse-discovering/). Except where otherwise noted, content on this site is licensed under the CC-BY license.
🔗 Finding the right tool for the job
The hardest part about effectively using R is finding the best packages for the problem you are trying to solve. I think this is even more important than being fluent in the language itself, which you will pick up along the way as you start using R more frequently. However, building your code on reliable foundations is essential for good results, and difficult to fix later on in a project.
There are about 20.000 packages on CRAN, and more on other networks such as BioConductor and GitHub. New packages are released every day. The developers are a mix of volunteers, ranging from professional software engineers to students and hobbyists. The quality and scope of packages varies enormously, which can make it difficult to judge which tools are the best choice for a given task.
The ultimate goal of r-universe is to help you effectively navigate the R ecosystem to discover what is out there, get a sense of the purpose and quality of individual packages, their developers, and get started using packages immediately and without any hassle.
🔗 Three levels of navigation in r-universe
We can distinguish three levels of navigation in r-universe when you go shopping for R packages:
- Search the global ecosystem: find packages, by topic, keyword, ranking, etc.
- Browse by maintainer/organization: explore all work from a given group or researcher.
- The individual package: get detailed information on everything there is to know about a project and instructions to start using it.
If you are new to R or are exploring packages for a certain task, you may start by searching everything that is out there using the r-universe search engine. Once you become more familiar with the ecosystem, you may get interested to zoom in to a specific suite of packages from a given maintainer or organization, and eventually learn more about the details of particular packages.
🔗 Level 1: searching the entire R ecosystem
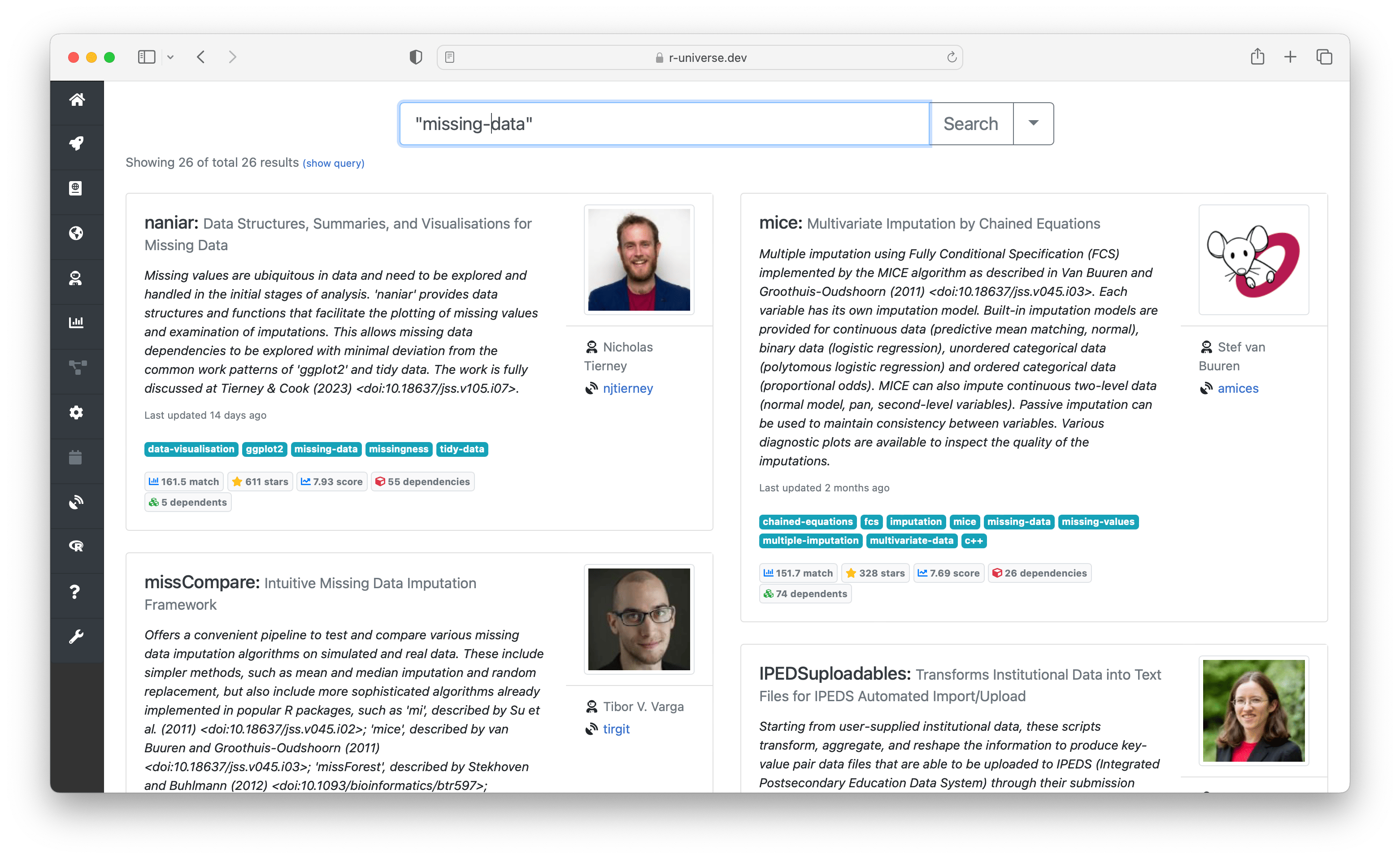
The landing page on https://r-universe.dev takes you directly to the global search engine. Here you can perform free-text search for a keyword, which is matched against package names, descriptions, topics, and titles of manual pages, vignettes, and so on. Results are ordered using a combination of a matching score (how often and where the search term appears), and a quality ranking1.
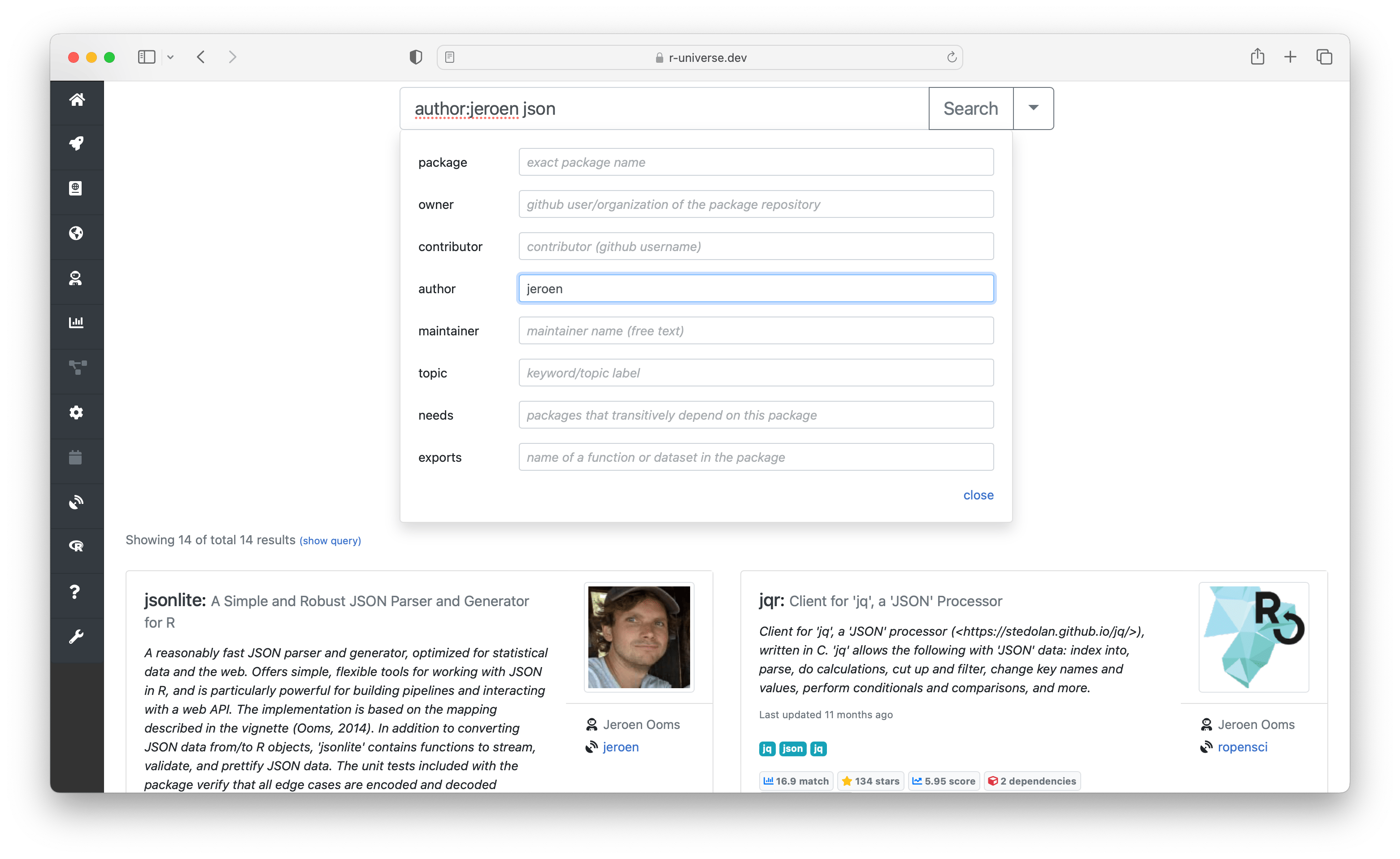
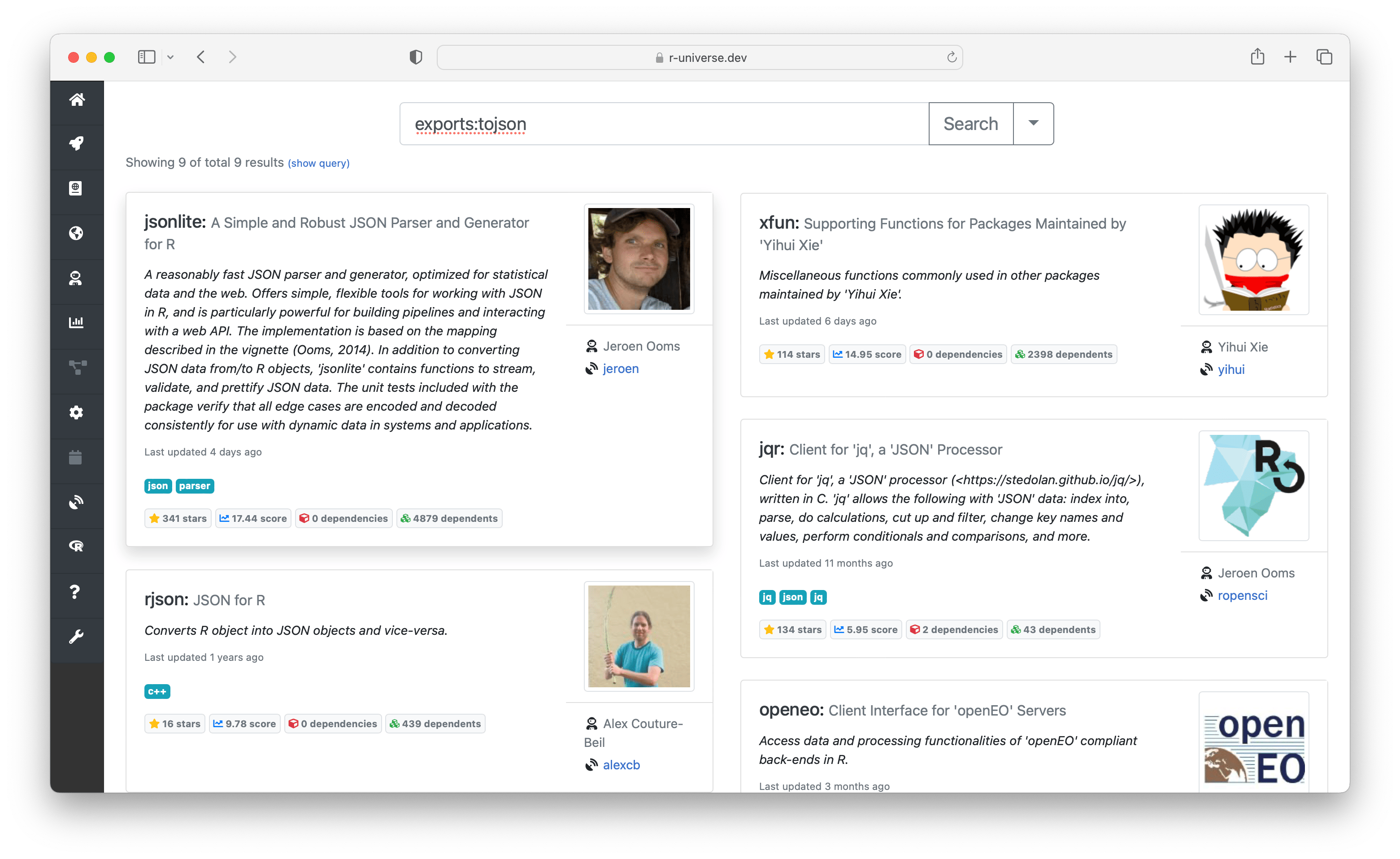
The right side of the search box contains an expand button which reveals powersearch filters. Here you can enter advanced search criteria, in order to specifically match specific fields. For example you can search for packages from a given author, or have a specific function or dependency.
The search page also shows a shuffling list of organizations that publish R packages (sorted by recent activity) which is a fun way to discover what is currently being developed in the R ecosystem.
🔗 Level 2: browsing work from a maintainer or organization
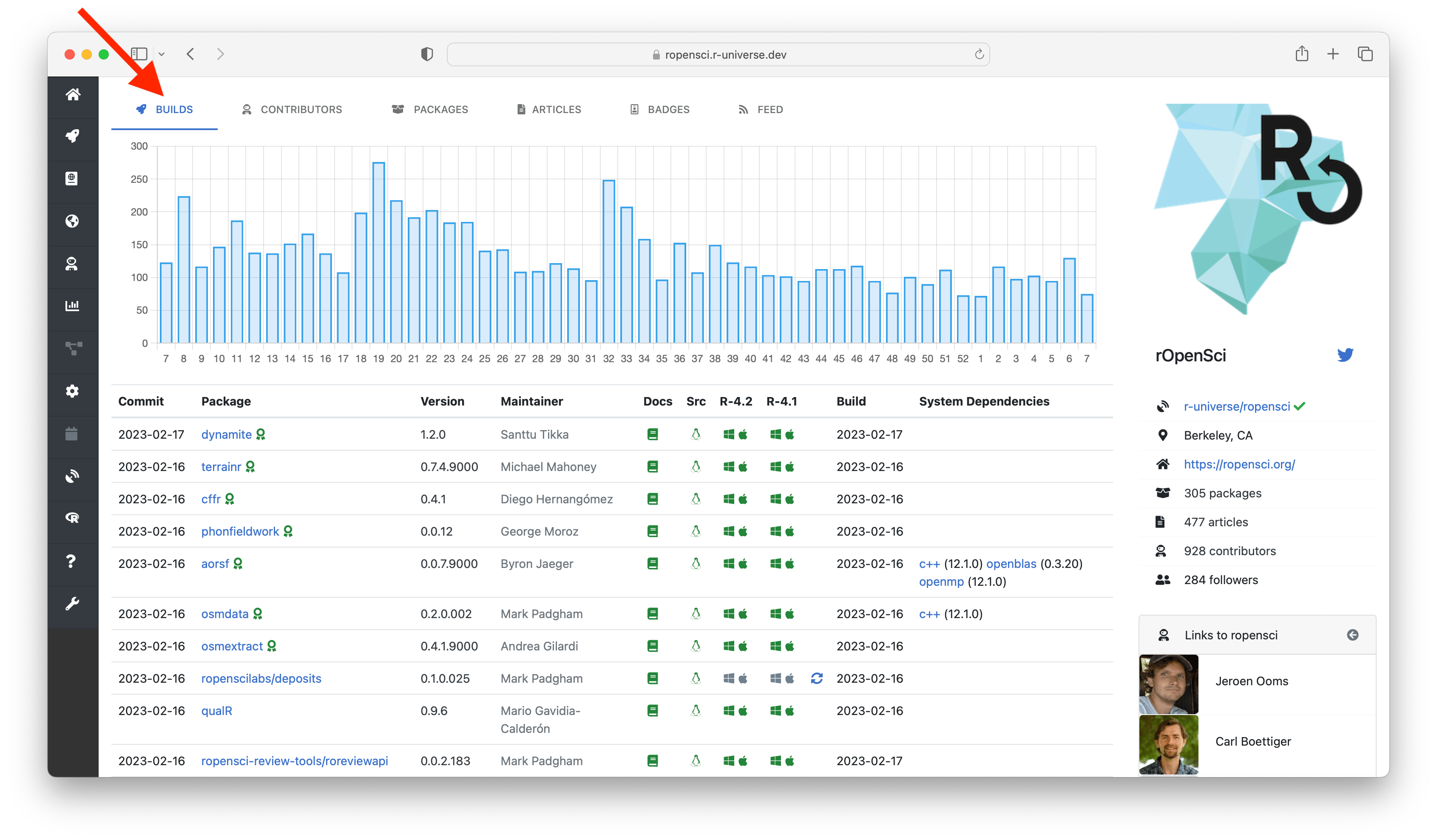
Each user and organization in r-universe has a unique subdomain under r-universe.dev with a personal cran-like package repository from where packages can directly be installed. For example, the packages and information from rOpenSci can be found on https://ropensci.r-universe.dev, and packages maintained by me are listed at https://jeroen.r-universe.dev (note how these two sets overlap).
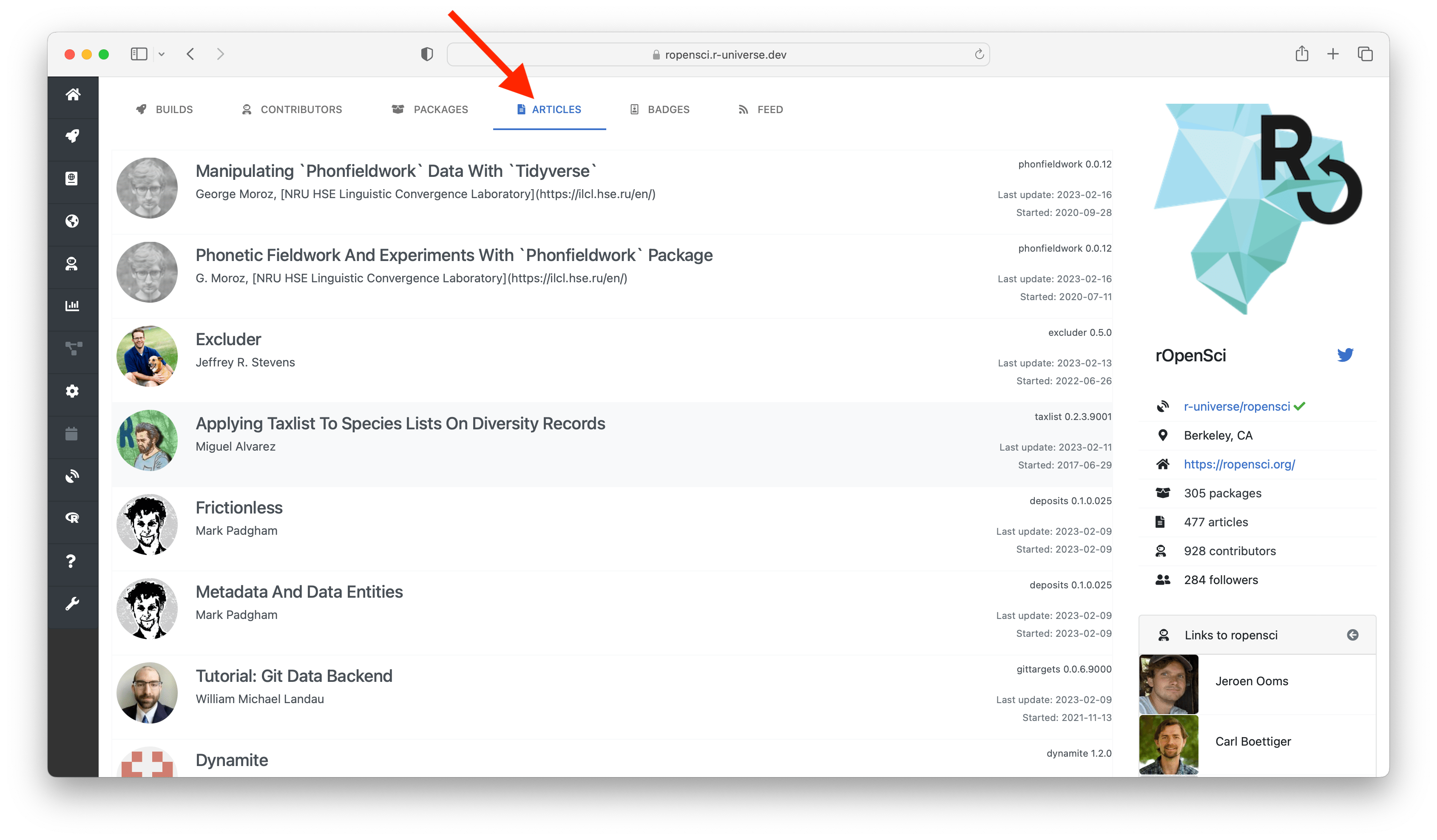
The top tabs allow you to explore different resources within universe. The builds tab shows the current state of the repository: a table with the name and version of each package, including the commit date, author, and build status. Clicking on the package name in the table sends you to the homepage of the package (more on this below), and the maintainer name links to the subdomain of that maintainer, listing other projects maintained by this person.
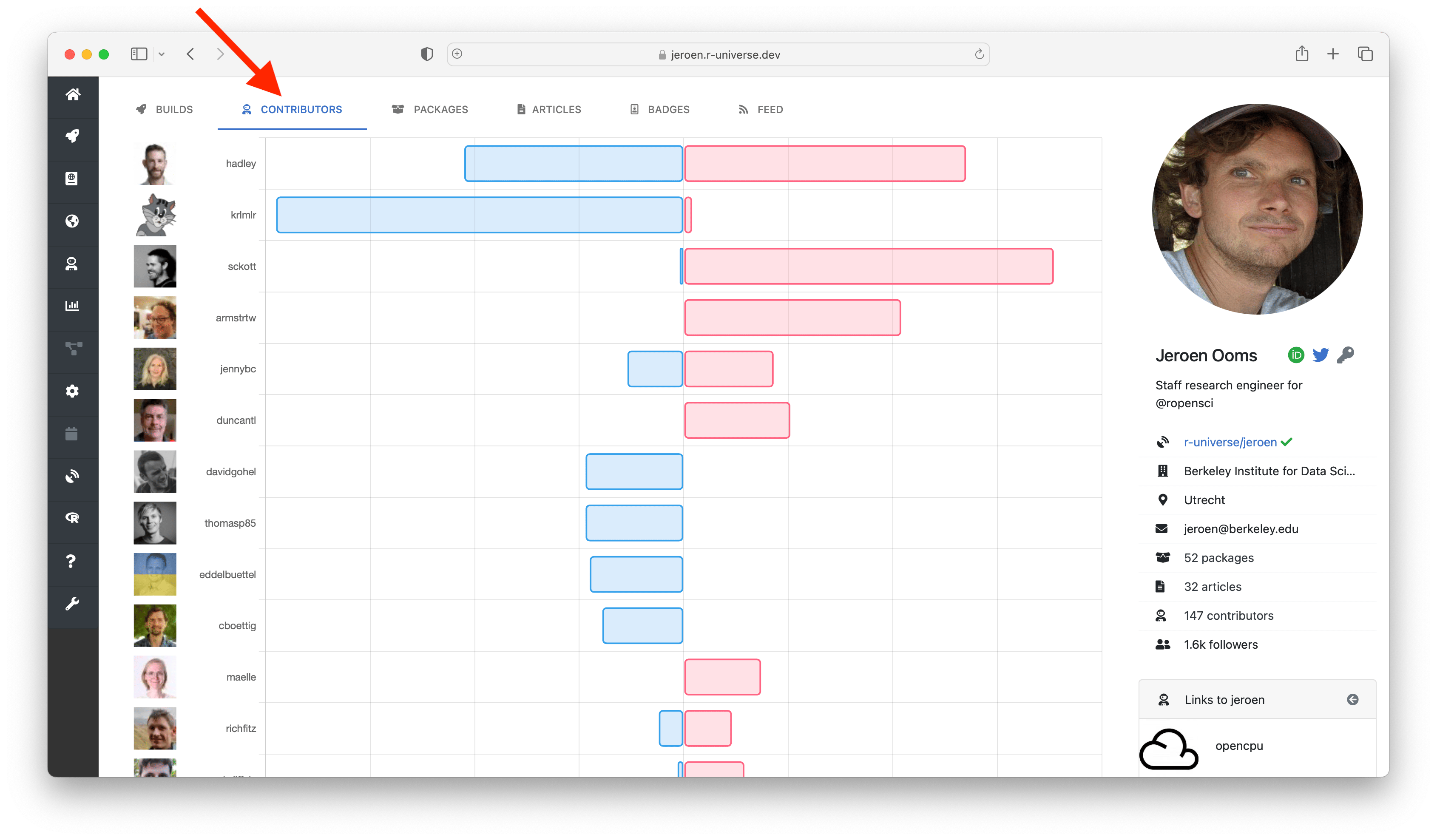
The packages tab lists descriptive information from all packages in the repository, and the articles tab shows all documents (e.g. vignettes). The contributors tab shows a chart with contributor statistics for the organization or maintainer. Clicking on the a bar in the chart sends you to this other universe where you can explore more work by this person.
The same subdomain hosts the actual cran-like package repository for this user or organization. Hence in order to list or install packages from rOpenSci in R, you would use:
# List available packages
available.packages(repos = "https://ropensci.r-universe.dev")
# Install a package
install.packages("magick", repos = "https://ropensci.r-universe.dev")
🔗 Level 3: individual package information
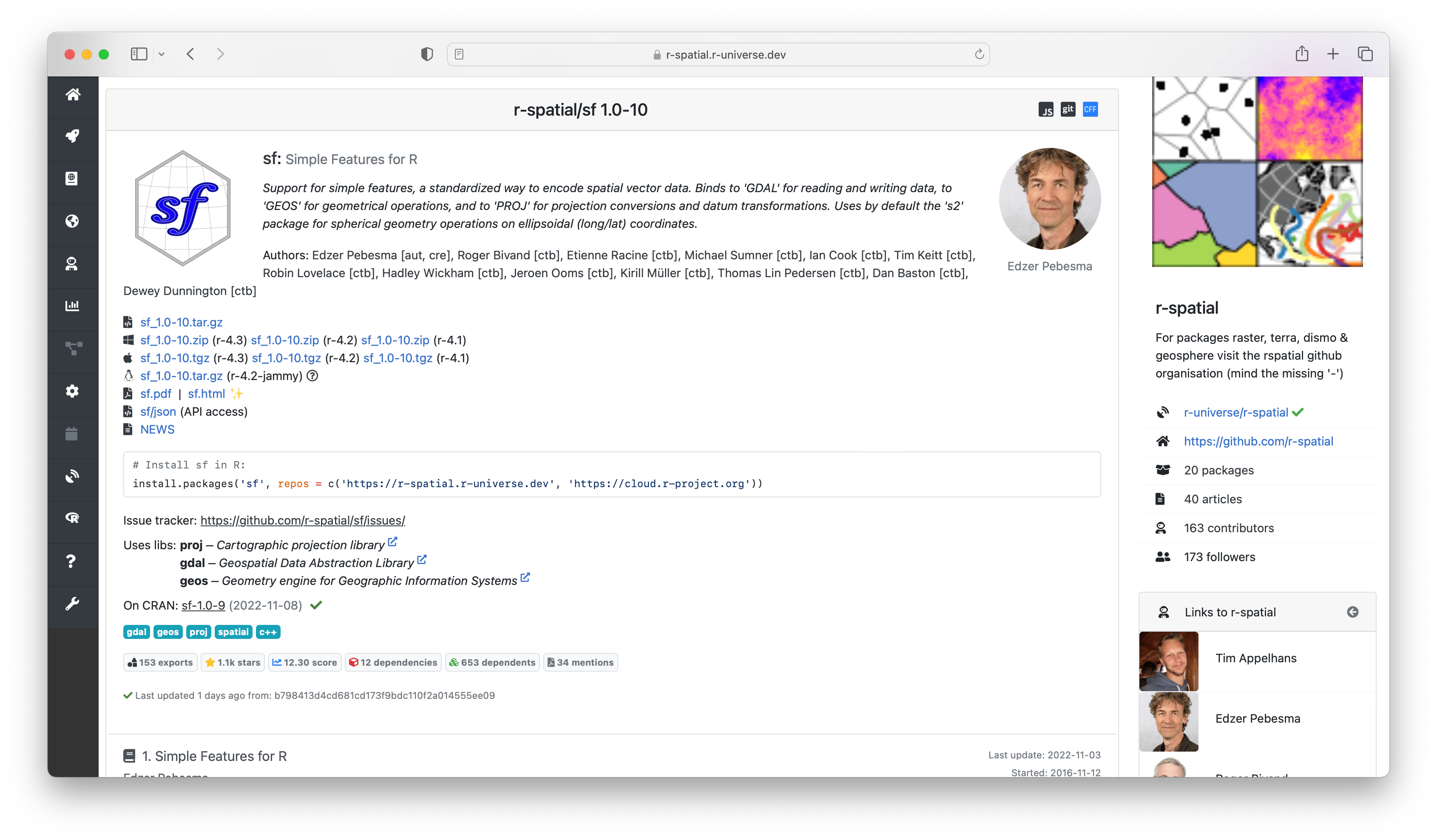
Finally we arrive at the individual package level. Clicking on a package in search results or in a universe overview will lead you to homepage for that package. You can also link to this page directly via the url https://{owner}.r-universe.dev/{package} for example https://r-spatial.r-universe.dev/sf below.
These package pages are generated automatically and provide detailed information about each project, and all the information and resources to quickly install and try the package locally in R.
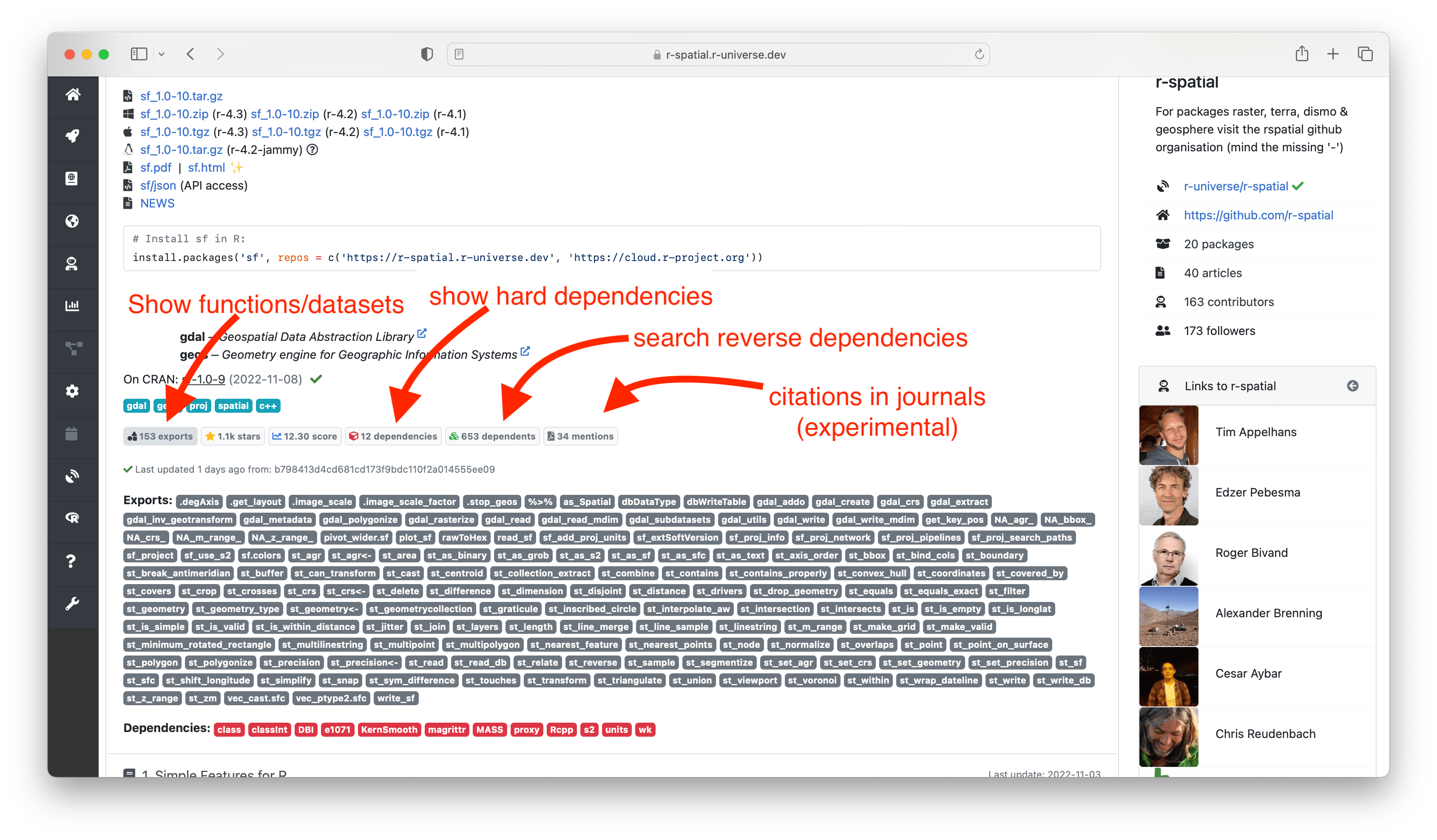
🔗 Package information
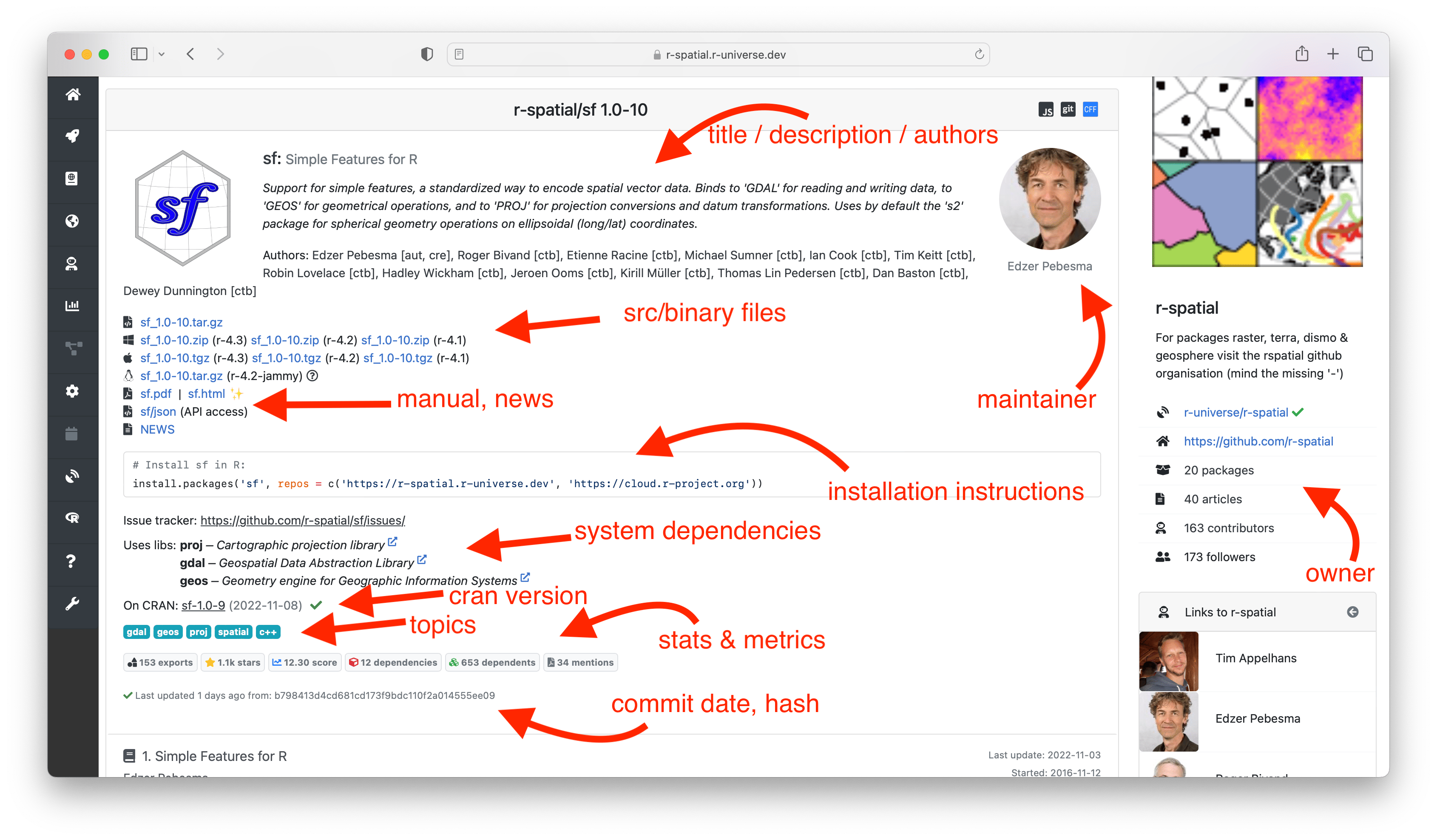
The initial section of the homepage shows descriptive data and links to resources from this package, such as source/binary files, the package manual and NEWS file. Further down we show installation instructions to install the package in R. Installation is quick and easy because we provide pre-compiled binaries for Windows and MacOS, so no special tools or setup is required on the user machine.
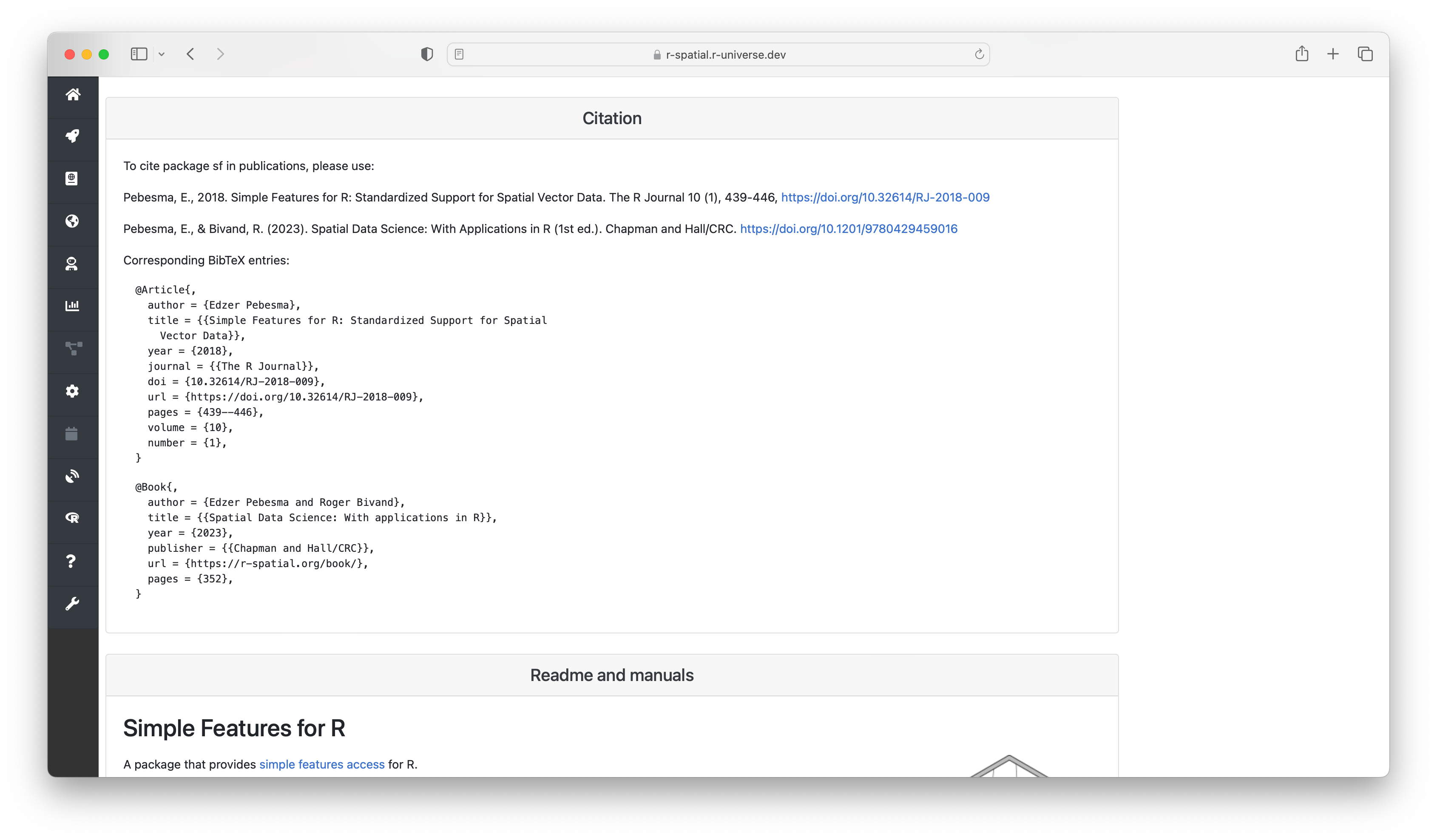
Here you can also find more information derived by the build system, such as which system libraries the package builds on, topics, and if the package is on CRAN. The badges underneath show statistics about the package, such as dependencies, reverse dependencies, and GitHub stars. Clicking on the ’exports’ badge will show all functions and datasets exported by the package, which link to the respective sections in the manual page. If specified, we also show how the package should be cited in papers.
🔗 All the documentation
To let you start using packages as quickly as possible, r-universe renders all available documentation and presents this as clear and concisely as possible on the homepage.
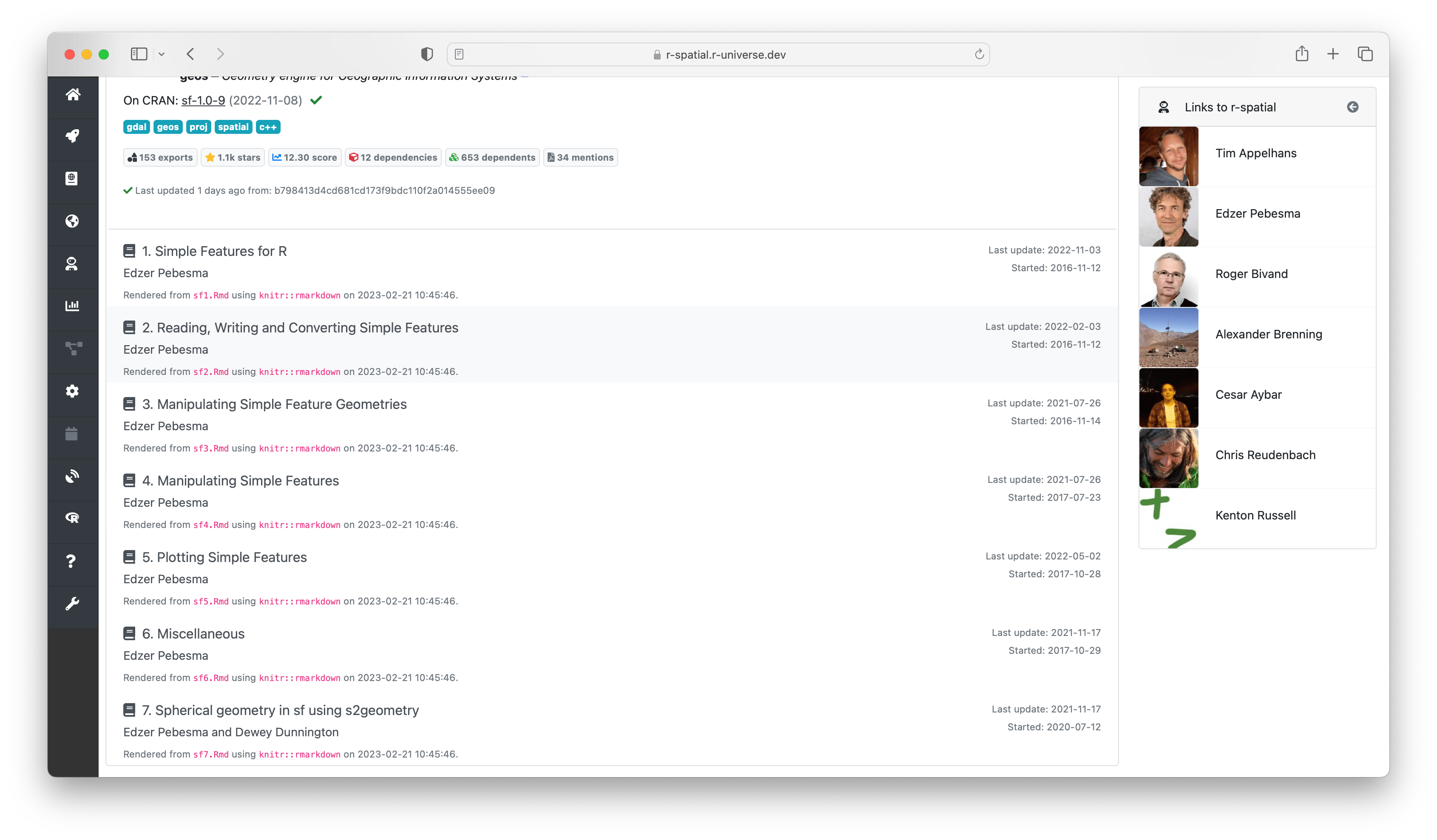
Most R packages have 3 forms of documentation: a readme file, a reference manual (help pages), and a number of articles (vignettes). These articles are often a good place to get started, and are listed on the package homepage right under the descriptives. From here you can view both the rendered html/pdf document, as well as the rmarkdown source file used to (re)produce the article.
Further down the page you can see the content from the package readme. Most packages these days include a README.md file which can nicely be rendered into HTML.
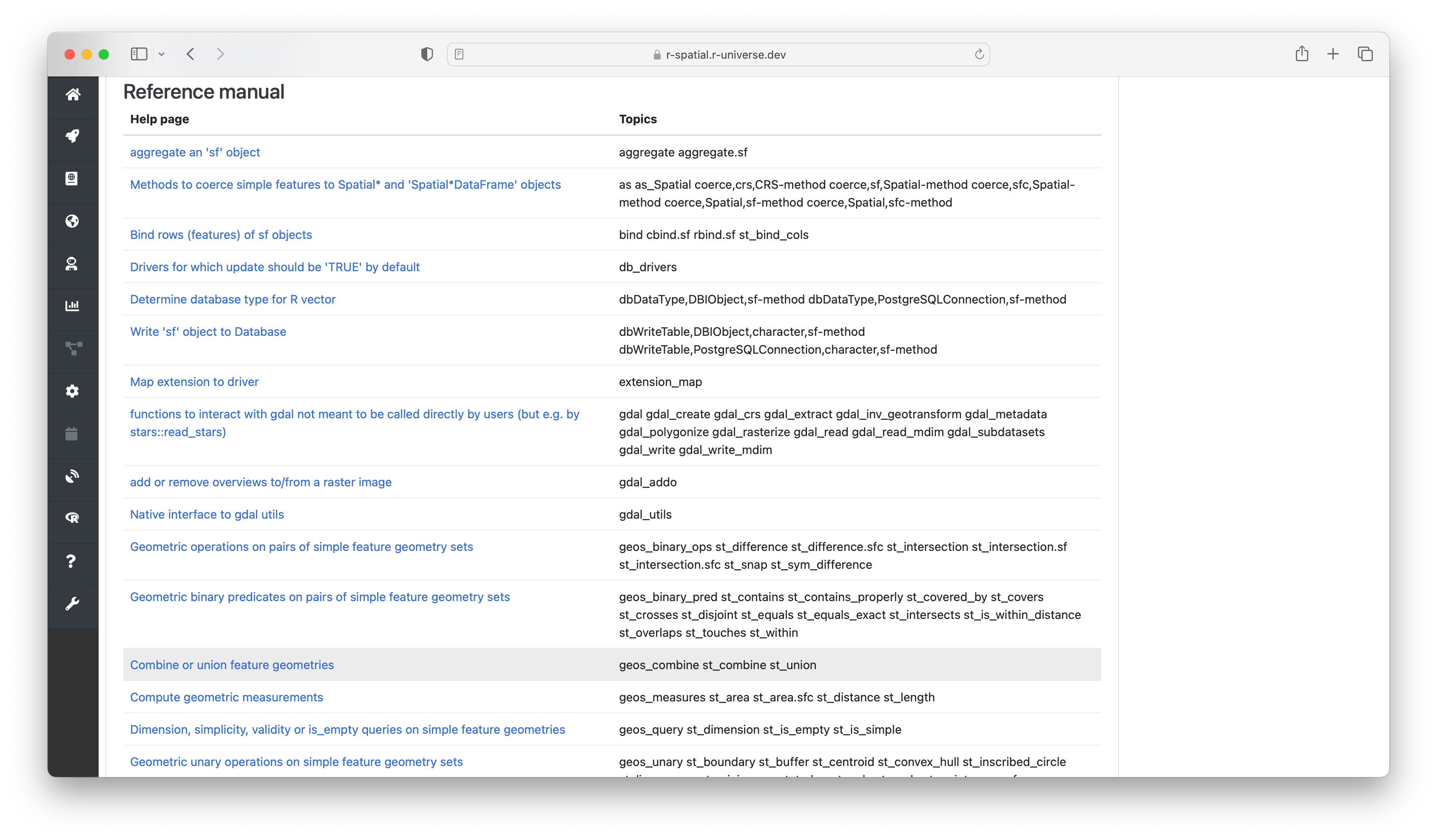
After the readme follows a section named “reference manual” containing a table with all the help pages. These link to the respective chapters in the html reference manual2, which provide the most extensive documentation for each function in the package, including descriptions, parameters, examples, etc.
🔗 Usage and development activity
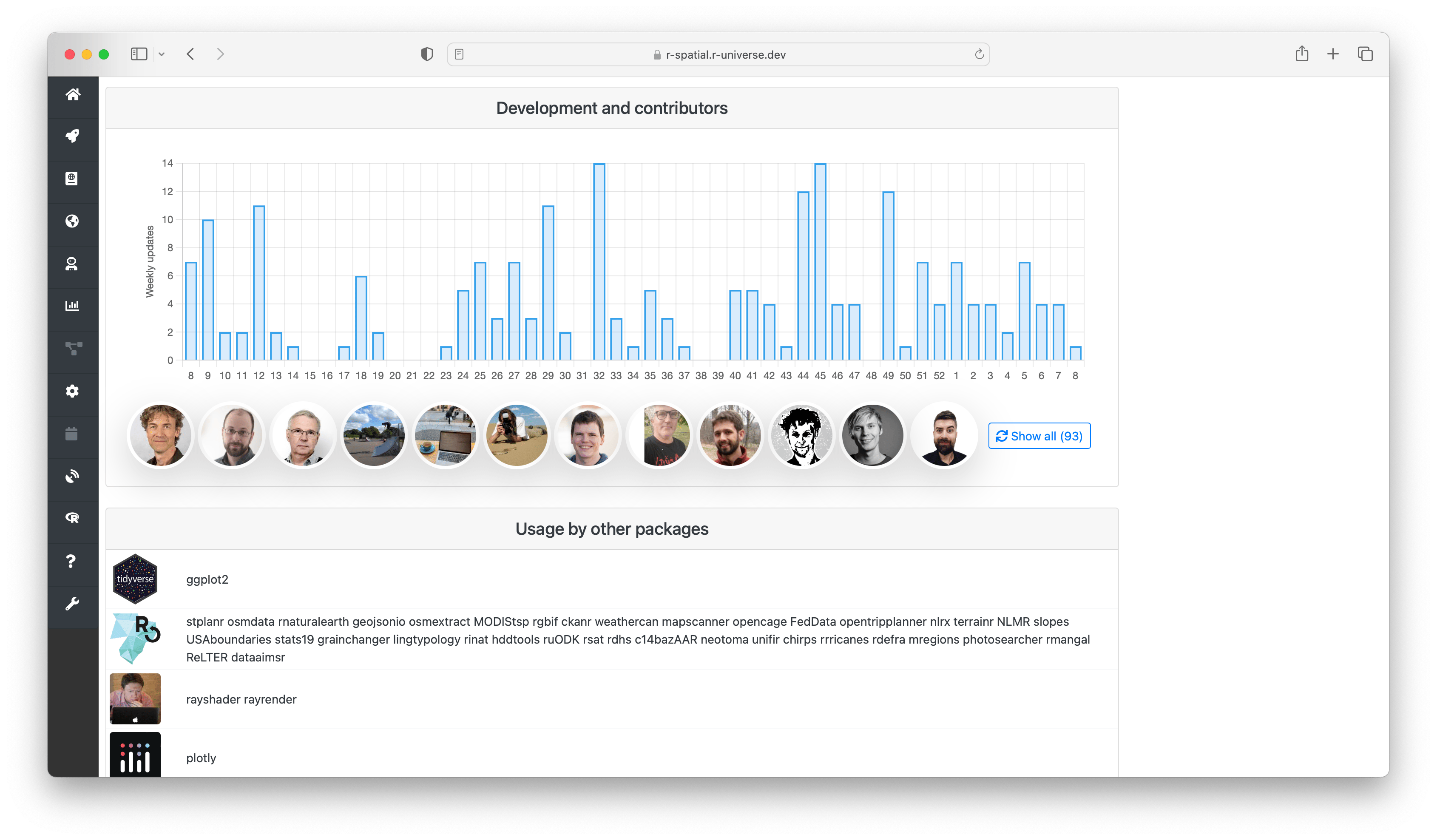
Besides familiarizing yourself with what the package does, you may be interested in who is working on it, who else is using it, and if it is still actively developed.
The “development and contributors” section shows a bar chart with the number of commits per week for the last year, and who the main contributors are. Finally the section “usage by other packages” shows other packages on r-universe depending on this package, grouped by owner. This gives you some indication on how active the project is, and who are involved.
🔗 Conclusion
This post explained different ways in which you can use r-universe to explore R packages. The search engine lets you find packages from anywhere in the ecosystem matching certain keywords or specific criteria. This is a good place to get stated and get a sense of what is available in a given domain.
Once you become more familiar with the ecosystem, you may get interested to zoom in to a specific suite of packages from a given group or researcher. The r-universe domain structure lets you browse packages and articles by organization, and discover related work with similar maintainers, contributors, or users.
Finally, for each R package, r-universe provides a beautiful homepage containing a wealth of information at: https://{owner}.r-universe.dev/{package}. This page shows exactly what a package is about, who is developing it, the status of the project, and also provides all nicely rendered documentation and installation instructions to start using the package immediately, without hassle.
More information about this ranking: how is the package-rank score calculated ↩︎
Blog post: postdoc 1.0: minimal and uncluttered HTML package manuals. ↩︎