All posts (Page 78 of 129)
Last week Google and friends released the new major version of their OCR system: Tesseract 4. This release builds upon 2+ years of hard work and has completely overhauled the internal OCR engine. From the tesseract wiki:
Tesseract 4.0 includes a new neural network-based recognition engine that delivers significantly higher accuracy (on document images) than the previous versions, in return for a significant increase in required compute power. On complex languages however, it may actually be faster than base Tesseract.
...
While many people groan at the thought of participating in a group ice breaker activity, we’ve gotten consistent feedback from people who have been to recent rOpenSci unconferences.
Best ice breaker ever!
We’ve had lots of requests for a detailed description of how we do it. This post shares our recipe, including a script you can adapt, a reflection on its success, examples of how others have used it, and some tips to remember. Let us know in the comments if you’ve used or adapted it!
...rOpenSci’s software engineer / postdoc Jeroen Ooms will explain what images are, under the hood, and showcase several rOpenSci packages that form a modern toolkit for working with images in R, including opencv, av, tesseract, magick and pdftools.
🕘 Thursday, November 15, 2018, 10-11AM PST; 7-8PM CET (find your timezone)
☎️ Find all details for joining the call on our Community Calls page. Everyone is welcome. No RSVP needed.
🔗
Agenda
- Welcome (Stefanie Butland, rOpenSci Community Manager, 5 min)
- Working with images in R (Jeroen Ooms, 35 min)
- Q & A (20 min)
🔗
Abstract
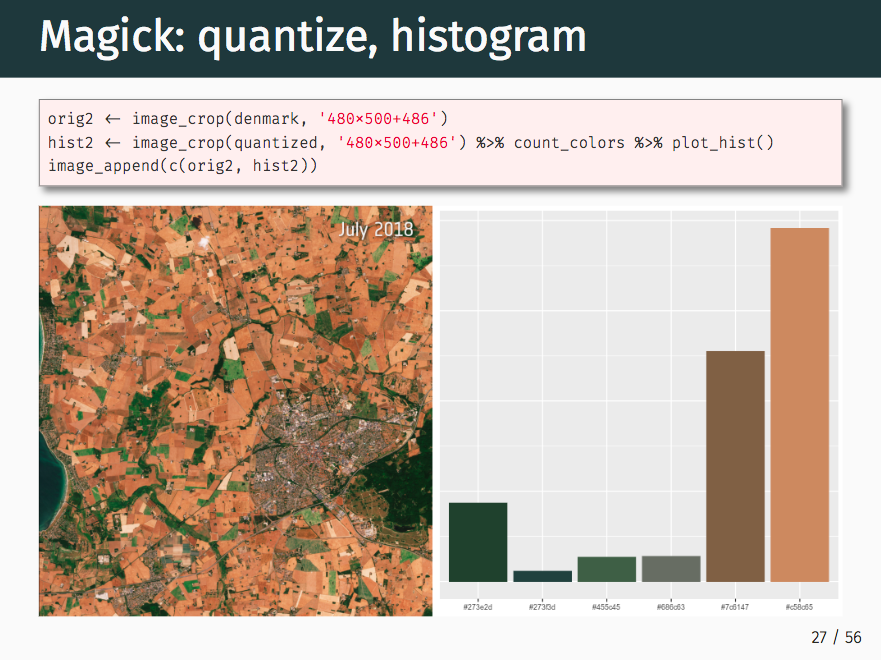
Images in various forms are used for numerous applications across scientific disciplines. Whether you are observing through satellite or microscope, looking at MRI scans or petri dishes, trying to find patterns or abnormalities, the data is in the image. Unfortunately the tools for working with images are traditionally highly fragmented by field, and often narrow in scope. At rOpenSci we are working on a suite of general purpose packages based on powerful c/c++ libraries. These provide an extensible and interoperable foundation for working with images in R, which can be used to implement domain specific-methods. This talk gives a taste of things we can currently do with images in R, and highlights some of the ongoing developments and challenges.
...pubchunks is a package grown out of the fulltext package. fulltext
provides a single interface to many sources of full text scholarly articles. As
part of the user flow in fulltext there is an extraction step where fulltext::chunks()
pulls parts of articles out of XML format article files.
As part of making fulltext more maintainable and focused on simply fetching articles,
and realizing that pulling out bits of structured XML files is a more general problem,
we broke out pubchunks into a separate package. fulltext::ft_chunks() and
fulltext::ft_tabularize() will eventually be removed and we’ll point users to
pubchunks.
Every R package has its story. Some packages are written by experts, some by
novices. Some are developed quickly, others were long in the making. This is the
story of jstor, a package which I developed during my time as a student of
sociology, working in a research project on the scientific elite within
sociology. Writing the package has taught me many things (more on that later)
and it is deeply gratifying to see, that others find the package useful....




