Wednesday, August 22, 2018 From rOpenSci (https://ropensci.org/blog/2018/08/22/rgbif-seven-years/). Except where otherwise noted, content on this site is licensed under the CC-BY license.
rgbif was seven years old yesterday!
🔗 What is rgbif?
rgbif gives you access to data from the Global Biodiversity Information Facility (GBIF) via their API.
A samping of use cases covered in rgbif:
- Search for datasets
- Get metrics on usage of datasets
- Get metadata about organizations providing data to GBIF
- Search taxonomic names
- Get quick taxonomic name suggestions
- Search occurrences by taxonomic name/country/collector/etc.
- Download occurrences by taxonomic name/country/collector/etc.
- Fetch raster maps to quickly visualize large scale biodiversity
🔗 History
Our first commit on rgbif was on 2011-08-26, uneventfully adding an empty README:
We’ve come a long way since Aug 2011. We’ve added a lot of new functionality and many new contributors.
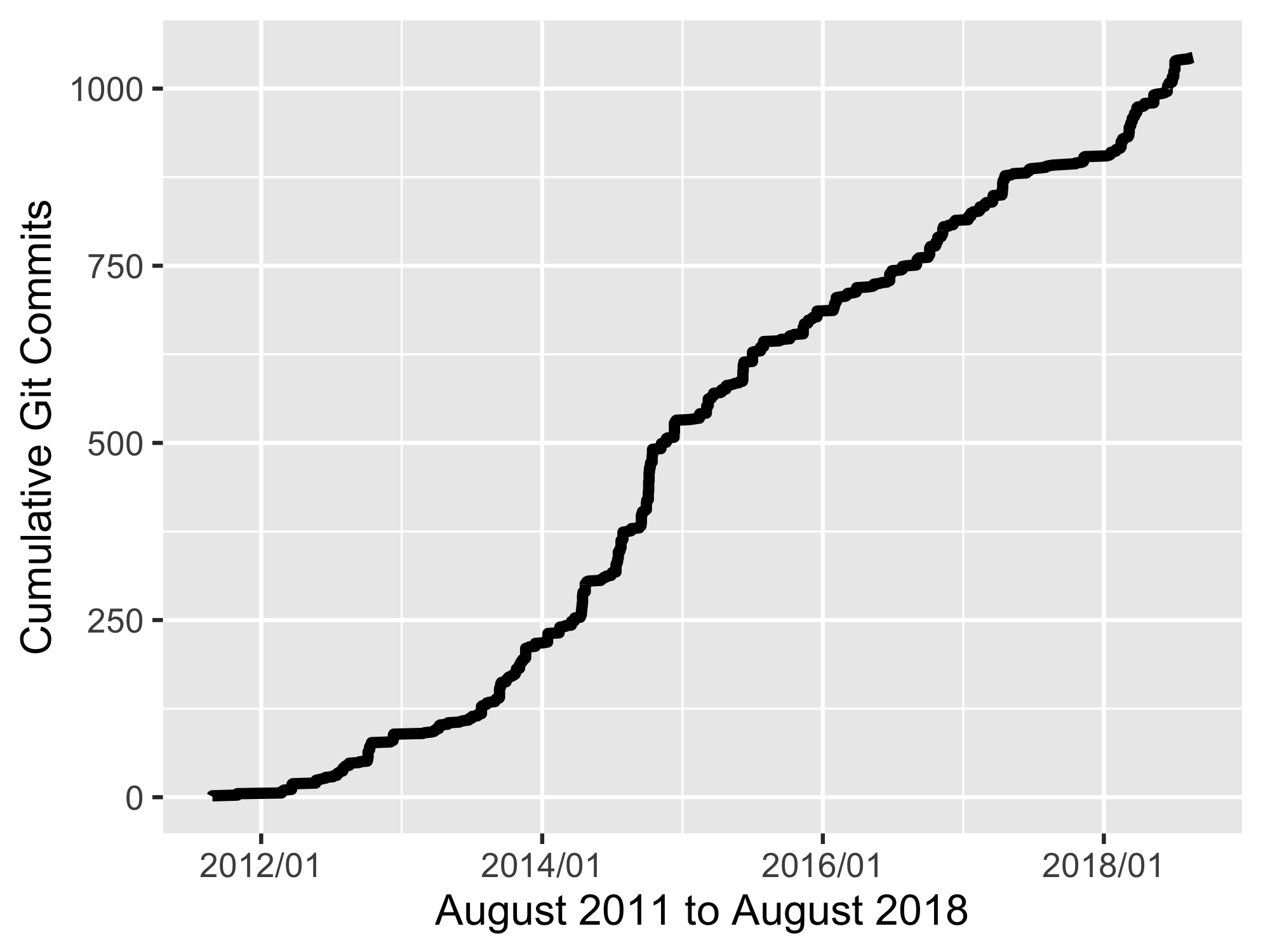
🔗 Commit history
Get git commits for rgbif using a few packages as well as git2r, our R package for working with git repositories:
library(git2r)
library(ggplot2)
library(dplyr)
repo <- git2r::repository("~/github/ropensci/rgbif")
res <- commits(repo)
A graph of commit history
dates <- vapply(res, function(z) {
as.character(as.POSIXct(z$author$when$time, origin = "1970-01-01"))
}, character(1))
df <- tbl_df(data.frame(date = dates, stringsAsFactors = FALSE)) %>%
group_by(date) %>%
summarise(count = n()) %>%
mutate(cumsum = cumsum(count)) %>%
ungroup()
ggplot(df, aes(x = as.Date(date), y = cumsum)) +
geom_line(size = 2) +
theme_grey(base_size = 16) +
scale_x_date(labels = scales::date_format("%Y/%m")) +
labs(x = 'August 2011 to August 2018', y = 'Cumulative Git Commits')
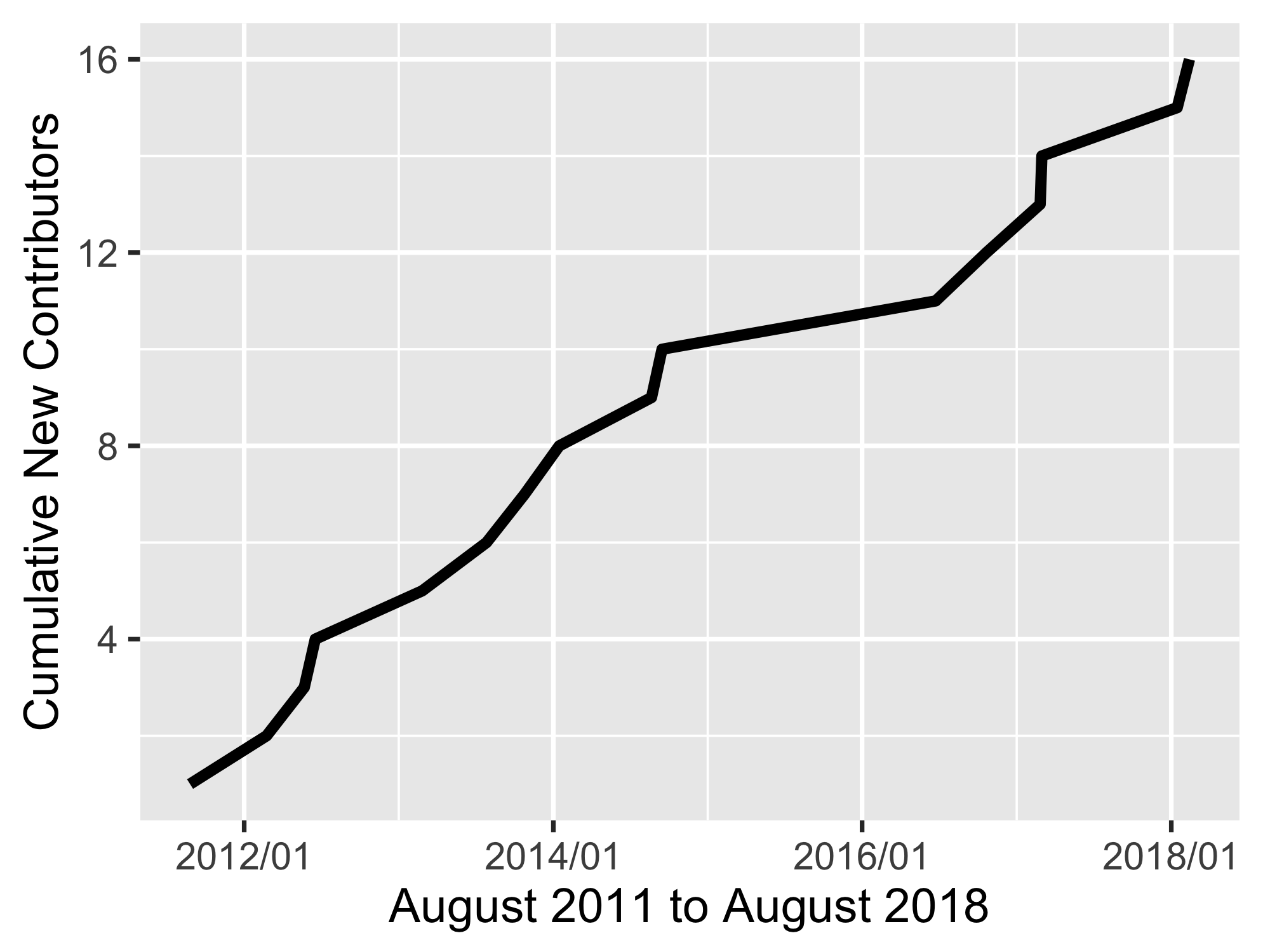
🔗 Contributors
A graph of new contributors through time
date_name <- lapply(res, function(z) {
data_frame(
date = as.character(as.POSIXct(z$author$when$time, origin = "1970-01-01")),
name = z$author$name
)
})
date_name <- bind_rows(date_name)
firstdates <- date_name %>%
group_by(name) %>%
arrange(date) %>%
filter(rank(date, ties.method = "first") == 1) %>%
ungroup() %>%
mutate(count = 1) %>%
arrange(date) %>%
mutate(cumsum = cumsum(count))
## plot
ggplot(firstdates, aes(as.Date(date), cumsum)) +
geom_line(size = 2) +
theme_grey(base_size = 18) +
scale_x_date(labels = scales::date_format("%Y/%m")) +
labs(x = 'August 2011 to August 2018', y = 'Cumulative New Contributors')
rgbif contributors, including those that have opened issues (click to go to their GitHub profile):
adamdsmith - AgustinCamacho - AlexPeap - andzandz11 - AugustT - benmarwick - cathynewman - cboettig - coyotree - damianooldoni - dandaman - djokester - dlebauer - dmcglinn - dnoesgaard - DupontCai - EDiLD - elgabbas - emhart - fxi - gkburada - hadley - ibartomeus - JanLauGe - jarioksa - jhpoelen - jkmccarthy - johnbaums - jwhalennds - karthik - kgturner - Kim1801 - ljuliusson - luisDVA - martinpfannkuchen - MattBlissett - MattOates - maxhenschell - Pakillo - peterdesmet - PhillRob - poldham - qgroom - raymondben - rossmounce - sacrevert - sckott - scottsfarley93 - SriramRamesh - steven2249 - stevenpbachman - stevensotelo - TomaszSuchan - Uzma-165 - vandit15 - vervis - vijaybarve - willgearty - zixuan75
🔗 rgbif usage
Carl Boettiger and I wrote a preprint paper describing rgbif in 2017, in PeerJ Preprints.
Chamberlain SA, Boettiger C. (2017) R Python, and Ruby clients for GBIF species occurrence data. PeerJ Preprints 5:e3304v1 https://doi.org/10.7287/peerj.preprints.3304v1
In that paper we also discuss Python (pygbif) and Ruby (gbifrb) GBIF clients. Check those out if you also sling Python or Ruby.
The paper above and/or the package have been cited 56 times over the past 7 years.
The way rgbif is used in research is most often in download occurrence data for a set of study species.
One example comes from the paper
Carvajal-Endara, S., Hendry, A. P., Emery, N. C., & Davies, T. J. (2017). Habitat filtering not dispersal limitation shapes oceanic island floras: species assembly of the Galápagos archipelago. Ecology Letters, 20(4), 495–504. https://doi.org/10.1111/ele.12753
Carvajal-Endara et al.
In another example (note the mention of removing certain records based on GBIF flags, check out rgbif::occ_issues to learn more)
Werner, G. D. A., Cornwell, W. K., Cornelissen, J. H. C., & Kiers, E. T. (2015). Evolutionary signals of symbiotic persistence in the legume–rhizobia mutualism. Proc Natl Acad Sci USA, 112(33), 10262–10269. https://doi.org/10.1073/pnas.1424030112
Werner et al.
🔗 Some features coming down the road
- Fully automated pagination across the package. Some functions have automated pagination (
occ_search/occ_data/allname_functions). So users don’t have to do manual pagination. - Improved
map_fetch()function. We just released this function in the last version, but it’s still early days and needs to improve a lot based on your feedback - Improved occurrence downloading queue: we rolled this out recently but just like
map_fetchit’s in its early days and definitely has many rough edges. Please let us know what you think!
🔗 Thanks!
We all owe a large debt of gratitude to GBIF for making an awesome resource for all those using their data, and to all the organizations/people that contribute data to GBIF.
A huge thanks goes to all rgbif users and contributors! It’s great to see how useful rgbif has been through the years, and we look forward to making it even better moving forward.